Search
- Page Path
- HOME > Search
- Calcium & bone metabolism
- MicroRNA-181a-5p Curbs Osteogenic Differentiation and Bone Formation Partially Through Impairing Runx1-Dependent Inhibition of AIF-1 Transcription
- Jingwei Liu, Xueying Chang, Daming Dong
- Endocrinol Metab. 2023;38(1):156-173. Published online January 6, 2023
- DOI: https://doi.org/10.3803/EnM.2022.1516
- 1,579 View
- 101 Download
- 2 Web of Science
- 1 Crossref
-
 Abstract
Abstract
 PDF
PDF Supplementary Material
Supplementary Material PubReader
PubReader  ePub
ePub - Background
Evidence has revealed the involvement of microRNAs (miRNAs) in modulating osteogenic differentiation, implying the promise of miRNA-based therapies for treating osteoporosis. This study investigated whether miR-181a-5p influences osteogenic differentiation and bone formation and aimed to establish the mechanisms in depth.
Methods
Clinical serum samples were obtained from osteoporosis patients, and MC3T3-E1 cells were treated with osteogenic induction medium (OIM) to induce osteogenic differentiation. miR-181a-5p-, Runt-related transcription factor 1 (Runx1)-, and/or allograft inflammatory factor-1 (AIF-1)-associated oligonucleotides or vectors were transfected into MC3T3-E1 cells to explore their function in relation to the number of calcified nodules, alkaline phosphatase (ALP) staining and activity, expression levels of osteogenesis-related proteins, and apoptosis. Luciferase activity, RNA immunoprecipitation, and chromatin immunoprecipitation assays were employed to validate the binding relationship between miR-181a-5p and Runx1, and the transcriptional regulatory relationship between Runx1 and AIF-1. Ovariectomy (OVX)-induced mice were injected with a miR-181a-5p antagonist for in vivo verification.
Results
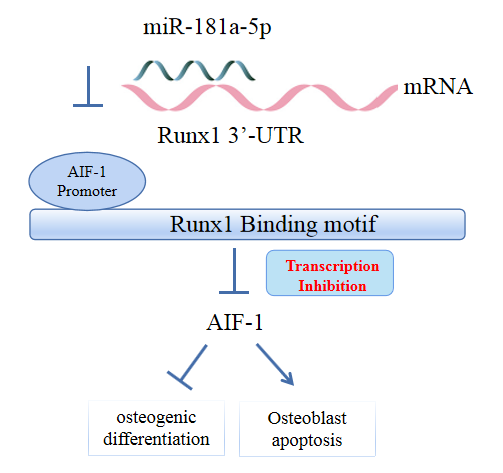
miR-181a-5p was highly expressed in the serum of osteoporosis patients. OIM treatment decreased miR-181a-5p and AIF-1 expression, but promoted Runx1 expression in MC3T-E1 cells. Meanwhile, upregulated miR-181a-5p suppressed OIM-induced increases in calcified nodules, ALP content, and osteogenesis-related protein expression. Mechanically, miR-181a-5p targeted Runx1, which acted as a transcription factor to negatively modulate AIF-1 expression. Downregulated Runx1 suppressed the miR-181a-5p inhibitor-mediated promotion of osteogenic differentiation, and downregulated AIF-1 reversed the miR-181a-5p mimic-induced inhibition of osteogenic differentiation. Tail vein injection of a miR-181a-5p antagonist induced bone formation in OVX-induced osteoporotic mice.
Conclusion
In conclusion, miR-181a-5p affects osteogenic differentiation and bone formation partially via the modulation of the Runx1/AIF-1 axis. -
Citations
Citations to this article as recorded by- Scopolamine regulates the osteogenic differentiation of human periodontal ligament stem cells through lactylation modification of RUNX2 protein
Ying Wu, Pan Gong
Pharmacology Research & Perspectives.2024;[Epub] CrossRef
- Scopolamine regulates the osteogenic differentiation of human periodontal ligament stem cells through lactylation modification of RUNX2 protein

- Diabetes, Obesity and Metabolism
- Inhibition of miR-146a-5p and miR-8114 in Insulin-Secreting Cells Contributes to the Protection of Melatonin against Stearic Acid-Induced Cellular Senescence by Targeting Mafa
- Shenghan Su, Qingrui Zhao, Lingfeng Dan, Yuqing Lin, Xuebei Li, Yunjin Zhang, Chunxiao Yang, Yimeng Dong, Xiaohan Li, Romano Regazzi, Changhao Sun, Xia Chu, Huimin Lu
- Endocrinol Metab. 2022;37(6):901-917. Published online December 7, 2022
- DOI: https://doi.org/10.3803/EnM.2022.1565

- 2,413 View
- 220 Download
- 2 Web of Science
- 2 Crossref
-
 Abstract
Abstract
 PDF
PDF Supplementary Material
Supplementary Material PubReader
PubReader  ePub
ePub - Background
Chronic exposure to elevated levels of saturated fatty acids results in pancreatic β-cell senescence. However, targets and effective agents for preventing stearic acid-induced β-cell senescence are still lacking. Although melatonin administration can protect β-cells against lipotoxicity through anti-senescence processes, the precise underlying mechanisms still need to be explored. Therefore, we investigated the anti-senescence effect of melatonin on stearic acid-treated mouse β-cells and elucidated the possible role of microRNAs in this process.
Methods
β-Cell senescence was identified by measuring the expression of senescence-related genes and senescence-associated β-galactosidase staining. Gain- and loss-of-function approaches were used to investigate the involvement of microRNAs in stearic acid-evoked β-cell senescence and dysfunction. Bioinformatics analyses and luciferase reporter activity assays were applied to predict the direct targets of microRNAs.
Results
Long-term exposure to a high concentration of stearic acid-induced senescence and upregulated miR-146a-5p and miR- 8114 expression in both mouse islets and β-TC6 cell lines. Melatonin effectively suppressed this process and reduced the levels of these two miRNAs. A remarkable reversibility of stearic acid-induced β-cell senescence and dysfunction was observed after silencing miR-146a-5p and miR-8114. Moreover, V-maf musculoaponeurotic fibrosarcoma oncogene homolog A (Mafa) was verified as a direct target of miR-146a-5p and miR-8114. Melatonin also significantly ameliorated senescence and dysfunction in miR-146a-5pand miR-8114-transfected β-cells.
Conclusion
These data demonstrate that melatonin protects against stearic acid-induced β-cell senescence by inhibiting miR-146a- 5p and miR-8114 and upregulating Mafa expression. This not only provides novel targets for preventing stearic acid-induced β-cell dysfunction, but also points to melatonin as a promising drug to combat type 2 diabetes progression. -
Citations
Citations to this article as recorded by- Genome-wide analysis in PC6 electroacupuncture to ameliorate carfilzomib-induced cardiotoxicity in mice
Yuxuan Chen, Rou Peng, Yi Qian, Yizhou Lu, Liyao Chen, Meiling Yu, Minjiao Jiang, Wei Wu, Shengfeng Lu
Gene.2024; 897: 148090. CrossRef - MiR-126 and miR-146a as Melatonin-Responsive Biomarkers for Neonatal Brain Ischemia
Maria Cristina Albertini, Tania Vanzolini, Serafina Perrone, Michael D. Weiss, Giuseppe Buonocore, Valentina Dell’Orto, Walter Balduini, Silvia Carloni
Journal of Molecular Neuroscience.2023; 73(9-10): 763. CrossRef
- Genome-wide analysis in PC6 electroacupuncture to ameliorate carfilzomib-induced cardiotoxicity in mice

- Obesity and Metabolism
- Role of PCSK9 Inhibitors in Patients with Familial Hypercholesterolemia
- Brian Tomlinson, Nivritti Gajanan Patil, Manson Fok, Christopher Wai Kei Lam
- Endocrinol Metab. 2021;36(2):279-295. Published online April 19, 2021
- DOI: https://doi.org/10.3803/EnM.2021.964

- 6,520 View
- 296 Download
- 14 Web of Science
- 15 Crossref
-
 Abstract
Abstract
 PDF
PDF PubReader
PubReader  ePub
ePub - Patients with familial hypercholesterolemia (FH) are at high or very high risk for cardiovascular disease. Those with heterozygous FH (HeFH) often do not reach low-density lipoprotein cholesterol (LDL-C) targets with statin and ezetimibe therapy, and those with homozygous FH (HoFH) usually require additional lipid-modifying therapies. Drugs that inhibit proprotein convertase subtilisin/kexin type 9 (PCSK9) offer a novel approach to reduce LDL-C. The monoclonal antibodies, alirocumab and evolocumab, given by subcutaneous injection every 2 or 4 weeks produce reductions in LDL-C of 50% to 60% in patients with HeFH, allowing many of them to achieve their LDL-C goals. Patients with HoFH show a reduced and more variable LDL-C response, which appears to depend on residual LDL receptor activity, and those with receptor-negative mutations may show no response. Inclisiran is a long-acting small interfering RNA therapeutic agent that inhibits the synthesis of PCSK9. Subcutaneous doses of 300 mg can reduce LDL-C by more than 50% for at least 6 months and the responses in HeFH and HoFH patients are similar to those achieved with monoclonal antibodies. These PCSK9 inhibitors are generally well tolerated and they provide a new opportunity for effective treatment for the majority of patients with FH.
-
Citations
Citations to this article as recorded by- Phenotypic homozygous familial hypercholesterolemia successfully treated with proprotein convertase subtilisin/kexin type 9 inhibitors
Ryosuke Tani, Keiji Matsunaga, Yuta Toda, Tomoko Inoue, Hai Ying Fu, Tetsuo Minamino
Clinical Case Reports.2024;[Epub] CrossRef - Targeting Lipoprotein(a): Can RNA Therapeutics Provide the Next Step in the Prevention of Cardiovascular Disease?
Henriette Thau, Sebastian Neuber, Maximilian Y. Emmert, Timo Z. Nazari-Shafti
Cardiology and Therapy.2024; 13(1): 39. CrossRef - Technologies of gene editing and related clinical trials for the treatment of genetic and acquired diseases: a systematic review
Wessam Sharaf-Eldin
Egyptian Journal of Medical Human Genetics.2024;[Epub] CrossRef - Qualitative and Quantitative Effects of PCSK9 Inhibitors in familial Hypercholesterolemia: a Synthetic Review
Aamina Shakir, Kyle Barron, Kalgi Modi
Current Problems in Cardiology.2023; 48(4): 101550. CrossRef - Inhibition of PCSK9 Improves the Development of Pulmonary Arterial Hypertension Via Down-Regulating Notch3 Expression
Peng Ye, Xiao-Min Jiang, Wei-Chun Qian, Juan Zhang
Cardiovascular Drugs and Therapy.2023;[Epub] CrossRef - Barriers and shortcomings in access to cardiovascular management and prevention for familial hypercholesterolemia during the COVID‐19 pandemic
Helen Huang, Keith S. K. Leung, Tulika Garg, Adele Mazzoleni, Goshen D. Miteu, Farida Zakariya, Wireko A. Awuah, Elaine T. S. Yin, Faaraea Haroon, Zarish Hussain, Narjiss Aji, Vikash Jaiswal, Gary Tse
Clinical Cardiology.2023; 46(8): 831. CrossRef - Familial Hypercholesterolemia in Children. The Current State of the Problem
Dinara I. Sadykova, Karina R. Salakhova, Liliya F. Galimova, Eugeniya S. Slastnikova, Chulpan D. Khaliullina
Current Pediatrics.2023; 22(3): 231. CrossRef - Long-term safety and effectiveness of alirocumab and evolocumab in familial hypercholesterolemia (FH) in Belgium
Marc Snel, Olivier S. Descamps
Acta Cardiologica.2023; : 1. CrossRef - PCSK9 inhibitors revisited: Effectiveness and safety of PCSK9 inhibitors in a real-life Spanish cohort
Juan Vicente-Valor, Xandra García-González, Sara Ibáñez-García, María Esther Durán-García, Ana de Lorenzo-Pinto, Carmen Rodríguez-González, Irene Méndez-Fernández, Juan Carlos Percovich-Hualpa, Ana Herranz-Alonso, María Sanjurjo-Sáez
Biomedicine & Pharmacotherapy.2022; 146: 112519. CrossRef - Development of small-molecule PCSK9 inhibitors for the treatment of hypercholesterolemia
Shakir Ahamad, Shintu Mathew, Waqas A. Khan, Kishor Mohanan
Drug Discovery Today.2022; 27(5): 1332. CrossRef - The biological relevance of PCSK9: when less is better…
Majambu Mbikay, Michel Chrétien
Biochemistry and Cell Biology.2022; 100(3): 189. CrossRef - Fenofibrate add-on to statin treatment is associated with low all-cause death and cardiovascular disease in the general population with high triglyceride levels
Kyung-Soo Kim, Sangmo Hong, Kyungdo Han, Cheol-Young Park
Metabolism.2022; 137: 155327. CrossRef - Homozygous Familial Hypercholesterolemia
Lisa Young, Emily E. Brown, Seth S. Martin
JACC: Case Reports.2022; 4(23): 101666. CrossRef - Familial Hypercholesterolemia and Its Current Diagnostics and Treatment Possibilities: A Literature Analysis
Kristina Zubielienė, Gintarė Valterytė, Neda Jonaitienė, Diana Žaliaduonytė, Vytautas Zabiela
Medicina.2022; 58(11): 1665. CrossRef - Efficacy and Safety of Alirocumab in Children and Adolescents With Homozygous Familial Hypercholesterolemia: Phase 3, Multinational Open-Label Study
Eric Bruckert, Sonia Caprio, Albert Wiegman, Min-Ji Charng, Cézar A. Zárate-Morales, Marie T. Baccara-Dinet, Garen Manvelian, Anne Ourliac, Michel Scemama, Stephen R. Daniels
Arteriosclerosis, Thrombosis, and Vascular Biology.2022; 42(12): 1447. CrossRef
- Phenotypic homozygous familial hypercholesterolemia successfully treated with proprotein convertase subtilisin/kexin type 9 inhibitors

- Effect of Dexamethasone and Deflazacort on the Function and Gene Expression of the Primary Cultured Human Osteoblast-Like Cells.
- Hyun Koo Yoon, In Myung Yang, Sung Woon Kim, Soung Seol Kim, Young Kil Choi, Ho Yeon Chung, Young Soon Kang, In Gul Moon, Chang Hoon Yim, Sang Woo Kim, Ki Ok Han, Hak Chul Chang, In Kwon Han
- J Korean Endocr Soc. 1996;11(4):479-491. Published online November 7, 2019
- 1,182 View
- 32 Download
-
 Abstract
Abstract
 PDF
PDF - Background
Chronic use of glucocorticoid is known to result in osteoporosis. Deflazacort (DFZ), a synthetic glucocorticoid, has been reported to have bone sparing properties in vivo eompared to dexamethasone(DEX). Not only the direct effect of DFZ on human osteoblast but the mechanism by which the drug spares bone remains unclear. This study, therefore, is aimed to investigate the direct effect of DFZ on the proliferation and differentiation of human osteoblast as well as on the gene expression of osteocalcin and osteoblast as well as on the gene expression of osteocalcin and growth factor produced in osteoblast. Methods: Human osteoblast-like cells were cultured from a piece of the tibia removed during selective orthopedic surgery for patients without metabolic bone diseases. The morphological iden- tification of osteoblast-like cell was performed under the light microscope after alkaline phosphatase staining. Cell proliferation rate was determined by [3H] thymidine incorporation into DNA. Cell differentiation was determined by alkaline phophatase activity. mRNA expression was quanti- tatively measured by the competitive reverse transcription-polymerase ehain reaction(RT-PCR). Results: The cultured cells demonstrated 1,25-dihydroxyvitamin D3-induced increases in alkaline phophatase activity and osteocalcin mRNA expression which are the properties of osteoblast. Twenty six percent of the cultured cells were identified as osteoblast-like cells by alkaline phophatase staining. After 24hr incubation with DEX or DFZ, the [3H) thymidine incorporation was significantly inhibited by 100nM DEX or DFL Alkaine phophatase activity was significantly increased by 100nM DEX. Osteocalcin mRNA was significantly decreased by both glueocorticoids. While DEX significantly suppressed expression of asteocalcin mRNA at 10nM and 100nM, DFZ did so only at 100nM. IGF-I mRNA was significantly decreased by 100nM DEX. Conclusion: These results suggest that the inhibitory effect of DFZ on the cell proliferation and protein synthesis is less than that of DEX, which might be responsible for the bone sparing effect of DFZ in vivo.

- Expression of Exon 1 and 6 of Indulin-like Growth Factor - 1 (IGF - 1) Gene in Thyroid Tissues.
- Sung Woon Kim, Hyun Ha Chang, In Kyung Jung, Jeong Taek Woo, In Myung Yang, Jin Woo Kim, Soung Seol Kim, Young Kil Choi
- J Korean Endocr Soc. 1996;11(4):409-417. Published online November 7, 2019
- 1,148 View
- 20 Download
-
 Abstract
Abstract
 PDF
PDF - Background
Goiter has been a common problem in the thyroid disease. The exact mechanism of goiter had not been clarified yet, but some goiters were increased with TSH(thyrotropin releasing hormone) dependent manner. TSH might be a major influencing factor for increasing size of goiter(goitrogen) and theres many cofactors those influenced to goiter size. One of the rnost prominent growth factor as a goitrogen is a IGF-I(insulin-like growth factor-I). IGF-I play a great role as a cofactor of goitrogen with TSH. This study, therefore, is aimed to investigate intracellular activation of IGF-I gene promoter in the surgical specimens of thyroid tumor. Methods: We used surgical specimen of various thyroid tissues from normal to malignant along its cell nature. Actually we used normal liver tissue as a IGF-I control tissue, normal thyroid, benign adenoma, and papillary thyroid cancer tissue with its malignat nature. We checked Mrna expression of whole IGF-I and IGF-I exon 6 by Northern blot method, and IGF-I, promoter 1 expression by RT-PCR-transcription method. Autoradiographied signals were analysed with densitometer. Results: We found whole IGF-I mRNAs were expressed with alternate splicing in exon 1, 2 and exon 4, 5 respectively. Striking events of IGF-I transcription were multiple tranascription initiatian in Pl and P2, and 3 sites for polyadenylation in exon 6. Four or more Mrna bands in Northern blot analysis of IGF-I(0.8, 1.4, 4.2, and 7.8kb) were noted. In low molecular weight IGF-I Mrna did not change their signal intensity with tissues, but exan 6(7.8kb) signals were significantly increased to its hepatic expression levels in malignant tissue. IGF-I, exon 1 expression by RT-PCR-T7 transcription was strikingly increased in thyroid cancer tissue, but exon 6 expression was not a great expession. Conclusion: One possible guess for this expression discrepancy of each exon may be originated from different Mrna degradation of each IGF-I signals. We need more preeise experiment for Mrna degradation speed of IGF-I.

- Expression of Epidermal Growth Factor Receptor mRNA by In Situ Hybridization in Normal and Abnormal Thyroid Tissue.
- Hyun Sik Son, Kun Ho Yoon, Bong Yun Cha, Jong Min Lee, Kwang Woo Lee, Moo Il Kang, Ho Young Son, Sung Koo Kang, Se Jeong Oh, Jin Han Kang, An Hee Lee
- J Korean Endocr Soc. 1994;9(4):337-343. Published online November 6, 2019
- 1,130 View
- 20 Download
-
 Abstract
Abstract
 PDF
PDF - Growth factors are polypeptide molecules that regulate cell growth and function by binding with high affinity to specific receptor molecules in the plasma membrane and stimulating receptor mediated action of intracellular signal transduction pathway.Epidermal growth factor(EGF) and their receptors(EGFR) regulate normal cellular growth, proliferation, and differentiation of various cells in vivo and in tissue cultures. And also may contribute directly to oncogenesis.Overexpression of EGFR and autocrine stimulation of growth involving this receptor system has been identified in several types of human neoplasia. There is evidence that the EGF and receptor system is involved in the regulation of follicular cell growth in the thyroid gland especially with immunohistochemical technic. But there was a challenge about the validity of previously performed immunohistochemical studies.In the study we investigated the relationship between EGFR mRNA expression and tumorigenesis by rapid in situ hybridization method. Formalin-fixed, paraffin embedded tissue sections of 10 normal, 17 nodular hyperplasia, 6 follicular adenoma, and 15 papillary cancer were examined. The results were as follows:1) EGFR mRNA positivity were 20%(2/10) in normal thyroid, 70%(12/17) in nodular hyperplasia, and 100% in follicular adenoma and papillary cancer.2) There was a significantly increased EGFR mRNA expression in papillary cancer compare to normal and nodular hyperplasia(p<0.05). But no difference was found with papillary cancer and follicular adenoma.3) There was a significantly increased EGFR mRNA expression in follicular adenoma compare to normal (p<0.05). But no difference was found with follicular adenoma and nodular hyperplasia. These results suggest that an overexpression of EGFR mRNA may play an important role in the tumorigenesis of thyroid tissue.

- In Situ Hybridization Analysis of Human Growth Hormone and Prolactin Secreting Pitultary Adenomas.
- Jae Wha Jo, Eun Jig Lee, Moon Suk Nam, Kyung Rae Kim, Sung Kil Lim, Hyun Chul Lee, Kap Bum Huh, Tae Seung Kim, Sun Ho Kim, Joong Uhn Choi, Kyu Chang Lee, Hyun Joo Jung, Sang Seop Chung
- J Korean Endocr Soc. 1994;9(2):82-92. Published online November 6, 2019
- 1,253 View
- 29 Download
-
 Abstract
Abstract
 PDF
PDF - A non-isotopic in situ hybridization method with biotin-labelled oligonucleotide probes was used to examine growth hormone(GH) and prolactin(PRL) gene expression in 32 patients with pituitary adenomas; 13 were prolactinomas, 8 GH secreting adenomas, and 11 mixed GH and PRL secreting adenomas.Positive immunostaining for GH was found in all patients with GH secreting adenomas, and mixed GH and PRL secreting adenomas. Positive immunostaining for PRL was found in all patients with prolactinomas and 9(81.8%) of 11 mixed GH and PRL secreting adenomas, 5(62.5%) of 8 GH secreting adenomas. Immunohistochemistry revealed that 13 were lactotrope adenomas, 5 somatotrope adenomas, and 14 GH and PRL cell adenomas.In situ hybridization revealed that GH mRNA expression was found in all the patients with somatotrope adenomas and GH and PRL cell adenomas, and 6(46.1%) of 13 lactotrope adenomas. PRL mRNA expression was 100% in lactotrope and GH and PRL cell adenomas, and 4(80.0%) of 5 somatotrope adenomas.The patients with a clinical diagnosis of acromegaly had detectable PRL mRNA in their neoplasm and it is suggested that the PRL cells in the adenomas did not result from dedifferentiation, but from the neoplastic stimulus for some mixed tumors probably occurred in cells previously committed to produce PRL and GH. In lactotrope adenomas, the PRL cells of the patients without expression of GH mRNA may be arised from cells programmed to secrete PRL or precussor PRL cells rather than from mixed GH-PRL cells. The finding that some patients produced mRNA detectable by in situ hybridization, but no hormone detectable by immunohistochemistry within tumor was suggested of a silent adenoma.These observations indicated that in situ hybridization studies may improve the classification of pituitary adenomas and may provide a precise knowledge of the biology of these neoplasms.

- Endocrine Research
- Transcriptome Analysis Identifies an Attenuated Local Immune Response in Invasive Nonfunctioning Pituitary Adenomas
- Yong Hwy Kim, Jung Hee Kim
- Endocrinol Metab. 2019;34(3):314-322. Published online September 26, 2019
- DOI: https://doi.org/10.3803/EnM.2019.34.3.314
- Correction in: Endocrinol Metab 2020;35(4):965
- 5,257 View
- 58 Download
- 12 Web of Science
- 10 Crossref
-
 Abstract
Abstract
 PDF
PDF Supplementary Material
Supplementary Material PubReader
PubReader  ePub
ePub Background Invasive nonfunctioning pituitary adenomas (NFPAs) remain challenging due to their high complication rate and poor prognosis. We aimed to identify the distinctive molecular signatures of invasive NFPAs, compared with noninvasive NFPAs, using gene expression profiling by RNA sequencing.
Methods We obtained frozen fresh tissue samples from 14 patients with NFPAs who underwent primary transsphenoidal surgery. Three non-invasive and 11 invasive NFPAs were used for RNA sequencing. The bioinformatics analysis included differential gene expression, gene ontology analysis, and pathway analysis.
Results A total of 700 genes were differentially expressed (59 up-regulated and 641 down-regulated genes) between invasive and non-invasive NFPAs (false discovery rate <0.1, and |fold change| ≥2). Using the down-regulated genes in invasive NFPAs, gene ontology enrichment analyses and pathway analyses demonstrated that the local immune response was attenuated and that transforming growth factor-β (TGF-β) RII-initiated TGF-β signaling was down-regulated in invasive NFPAs. The overexpression of claudin-9 (
CLDN9 ) and the down-regulation of insulin-like growth factor-binding protein 5 (IGFBP5 ), death-associated protein kinase 1 (DAPK1 ), and tissue inhibitor of metalloproteinase-3 (TIMP3 ) may be related with invasiveness in NFPAs.Conclusion Invasive NFPAs harbor different gene expression profiles relative to noninvasive NFPAs. In particular, local suppression of the immune response and TGF-β signaling can make PAs prone to invasiveness.
-
Citations
Citations to this article as recorded by- Transcriptome of GH-producing pituitary neuroendocrine tumours and models are significantly affected by somatostatin analogues
Rihards Saksis, Olesja Rogoza, Helvijs Niedra, Kaspars Megnis, Ilona Mandrika, Inga Balcere, Liva Steina, Janis Stukens, Austra Breiksa, Jurijs Nazarovs, Jelizaveta Sokolovska, Ilze Konrade, Raitis Peculis, Vita Rovite
Cancer Cell International.2023;[Epub] CrossRef - PD-L1 Expression in Pituitary Neuroendocrine Tumors/Pituitary Adenomas
Giulia Cossu, Stefano La Rosa, Jean Philippe Brouland, Nelly Pitteloud, Ethan Harel, Federico Santoni, Maxime Brunner, Roy Thomas Daniel, Mahmoud Messerer
Cancers.2023; 15(18): 4471. CrossRef - Aggressive PitNETs and Potential Target Therapies: A Systematic Review of Molecular and Genetic Pathways
Simona Serioli, Ludovico Agostini, Alberto Pietrantoni, Federico Valeri, Flavia Costanza, Sabrina Chiloiro, Barbara Buffoli, Amedeo Piazza, Pietro Luigi Poliani, Maria Peris-Celda, Federica Iavarone, Simona Gaudino, Marco Gessi, Giovanni Schinzari, Pier P
International Journal of Molecular Sciences.2023; 24(21): 15719. CrossRef - Muti-omics integration analysis revealed molecular network alterations in human nonfunctional pituitary neuroendocrine tumors in the framework of 3P medicine
Siqi Wen, Chunling Li, Xianquan Zhan
EPMA Journal.2022; 13(1): 9. CrossRef - A systematic review of molecular alterations in invasive non-functioning pituitary adenoma
Nazanin Hosseinkhan, Maryam Honardoost, Zahra Emami, Sara Cheraghi, Nahid Hashemi-Madani, Mohammad E. Khamseh
Endocrine.2022; 77(3): 500. CrossRef - Medication for Acromegaly Reduces Expression of MUC16, MACC1 and GRHL2 in Pituitary Neuroendocrine Tumour Tissue
Rihards Saksis, Ivars Silamikelis, Pola Laksa, Kaspars Megnis, Raitis Peculis, Ilona Mandrika, Olesja Rogoza, Ramona Petrovska, Inga Balcere, Ilze Konrade, Liva Steina, Janis Stukens, Austra Breiksa, Jurijs Nazarovs, Jelizaveta Sokolovska, Valdis Pirags,
Frontiers in Oncology.2021;[Epub] CrossRef - Large Scale Molecular Studies of Pituitary Neuroendocrine Tumors: Novel Markers, Mechanisms and Translational Perspectives
Raitis Peculis, Helvijs Niedra, Vita Rovite
Cancers.2021; 13(6): 1395. CrossRef - Nkx3-1 and Fech genes might be switch genes involved in pituitary non-functioning adenoma invasiveness
Nasibeh Khayer, Maryam Jalessi, Amin Jahanbakhshi, Alireza Tabib khooei, Mehdi Mirzaie
Scientific Reports.2021;[Epub] CrossRef - The tumour microenvironment of pituitary neuroendocrine tumours
Pedro Marques, Ashley B. Grossman, Márta Korbonits
Frontiers in Neuroendocrinology.2020; 58: 100852. CrossRef - The Progress of Immunotherapy in Refractory Pituitary Adenomas and Pituitary Carcinomas
Congxin Dai, Siyu Liang, Bowen Sun, Jun Kang
Frontiers in Endocrinology.2020;[Epub] CrossRef
- Transcriptome of GH-producing pituitary neuroendocrine tumours and models are significantly affected by somatostatin analogues

- Alternative Polyadenylation in Human Diseases
- Jae-Woong Chang, Hsin-Sung Yeh, Jeongsik Yong
- Endocrinol Metab. 2017;32(4):413-421. Published online December 14, 2017
- DOI: https://doi.org/10.3803/EnM.2017.32.4.413
- 5,445 View
- 94 Download
- 26 Web of Science
- 27 Crossref
-
 Abstract
Abstract
 PDF
PDF PubReader
PubReader Varying length of messenger RNA (mRNA) 3′-untranslated region is generated by alternating the usage of polyadenylation sites during pre-mRNA processing. It is prevalent through all eukaryotes and has emerged as a key mechanism for controlling gene expression. Alternative polyadenylation (APA) plays an important role for cell growth, proliferation, and differentiation. In this review, we discuss the functions of APA related with various physiological conditions including cellular metabolism, mRNA processing, and protein diversity in a variety of disease models. We also discuss the molecular mechanisms underlying APA regulation, such as variations in the concentration of mRNA processing factors and RNA-binding proteins, as well as global transcriptome changes under cellular signaling pathway.
-
Citations
Citations to this article as recorded by- PABPN1 loss-of-function causes APA-shift in oculopharyngeal muscular dystrophy
Milad Shademan, Hailiang Mei, Baziel van Engelen, Yavuz Ariyurek, Susan Kloet, Vered Raz
Human Genetics and Genomics Advances.2024; 5(2): 100269. CrossRef - Integration of genomic and transcriptomic data of inbred mouse models for polygenic obesity and leanness revealed “obese” and “lean” candidate alleles in polyadenylation signals
Martin Šimon, Špela Mikec, Nicholas M. Morton, Santosh S. Atanur, Simon Horvat, Tanja Kunej
Gene Reports.2024; 35: 101903. CrossRef - An AluYa5 Insertion in the 3′UTR of COL4A1 and Cerebral Small Vessel Disease
Chaker Aloui, Lisa Neumann, Françoise Bergametti, Eric Sartori, Marc Herbreteau, Arnaud Maillard, Thibault Coste, Hélène Morel, Dominique Hervé, Hugues Chabriat, Serge Timsit, Irina Viakhireva, Yves Denoyer, Rémi Allibert, Florence Demurger, Cedric Gollio
JAMA Network Open.2024; 7(4): e247034. CrossRef - Genome-wide screening for genetic variants in polyadenylation signal (PAS) sites in mouse selection lines for fatness and leanness
Martin Šimon, Špela Mikec, Nicholas M. Morton, Santosh S. Atanur, Janez Konc, Simon Horvat, Tanja Kunej
Mammalian Genome.2023; 34(1): 12. CrossRef - Alternative polyadenylation regulation in cardiac development and cardiovascular disease
Jun Cao, Muge N Kuyumcu-Martinez
Cardiovascular Research.2023; 119(6): 1324. CrossRef - Alternative polyadenylation reprogramming of MORC2 induced by NUDT21 loss promotes KIRC carcinogenesis
Yuqin Tan, Tong Zheng, Zijun Su, Min Chen, Suxiang Chen, Rui Zhang, Ruojiao Wang, Ke Li, Ning Na
JCI Insight.2023;[Epub] CrossRef - Emerging roles of alternative cleavage and polyadenylation (APA) in human disease
Prakash Dharmalingam, Rajasekaran Mahalingam, Hari Krishna Yalamanchili, Tingting Weng, Harry Karmouty‐Quintana, Ashrith Guha, Rajarajan A. Thandavarayan
Journal of Cellular Physiology.2022; 237(1): 149. CrossRef - Fe-S clusters masquerading as zinc finger proteins
Jordan D. Pritts, Sarah L.J. Michel
Journal of Inorganic Biochemistry.2022; 230: 111756. CrossRef - Alternative polyadenylation associated with prognosis and therapy in colorectal cancer
Yi Zhang, Yunfei Xu, Yuzhi Wang
Scientific Reports.2022;[Epub] CrossRef - Population‐scale genetic control of alternative polyadenylation and its association with human diseases
Lei Li, Yumei Li, Xudong Zou, Fuduan Peng, Ya Cui, Eric J. Wagner, Wei Li
Quantitative Biology.2022; 10(1): 44. CrossRef - Comprehensive analysis of alternative polyadenylation regulators concerning CD276 and immune infiltration in bladder cancer
Ming Xiong, Wencheng Li, Longwang Wang, Liang Chen, Zhaohui Chen, Chengcheng Wei, Futian Zhang, Jiawei Chen, Gallina Kazobinka, Jun Zhao, Teng Hou
BMC Cancer.2022;[Epub] CrossRef - Non-Allelic Homologous Recombination Leading to Premature Transcription Termination in the ARSB Gene as a Novel Cause of Mucopolysaccharidosis Type VI
Igor Bychkov, Filatova A. Yu, Galina V. Baydakova, Nataliya V. Sikora, Alexandr S. Skretnev, Tabakov V. Yu, Skoblov M. Yu, Zakharova E. Yu
SSRN Electronic Journal .2022;[Epub] CrossRef - Alternative polyadenylation of mRNA and its role in cancer
Fuwen Yuan, William Hankey, Eric J. Wagner, Wei Li, Qianben Wang
Genes & Diseases.2021; 8(1): 61. CrossRef - Animal-APAdb: a comprehensive animal alternative polyadenylation database
Weiwei Jin, Qizhao Zhu, Yanbo Yang, Wenqian Yang, Dongyang Wang, Jiajun Yang, Xiaohui Niu, Debing Yu, Jing Gong
Nucleic Acids Research.2021; 49(D1): D47. CrossRef - Emerging cellular and molecular determinants of idiopathic pulmonary fibrosis
Thị Hằng Giang Phan, Panagiotis Paliogiannis, Gheyath K. Nasrallah, Roberta Giordo, Ali Hussein Eid, Alessandro Giuseppe Fois, Angelo Zinellu, Arduino Aleksander Mangoni, Gianfranco Pintus
Cellular and Molecular Life Sciences.2021; 78(5): 2031. CrossRef - Alternative polyadenylation: methods, mechanism, function, and role in cancer
Yi Zhang, Lian Liu, Qiongzi Qiu, Qing Zhou, Jinwang Ding, Yan Lu, Pengyuan Liu
Journal of Experimental & Clinical Cancer Research.2021;[Epub] CrossRef - Understanding RNA Binding by the Nonclassical Zinc Finger Protein CPSF30, a Key Factor in Polyadenylation during Pre-mRNA Processing
Jordan D. Pritts, Abdulafeez A. Oluyadi, Weiliang Huang, Geoffrey D. Shimberg, Maureen A. Kane, Angela Wilks, Sarah L. J. Michel
Biochemistry.2021; 60(10): 780. CrossRef - Shortening of HO1 3′UTRs by Alternative Polyadenylation Suppresses Adipogenesis in 3T3-L1
Jianwei Cui, Chengping Li, Xiao Cui, Xueyan Liu, Chaoqun Meng, Guoli Zhou
Journal of Agricultural and Food Chemistry.2021; 69(28): 8038. CrossRef - The DEAD-box RNA helicase SHI2 functions in repression of salt-inducible genes and regulation of cold-inducible gene splicing
Bangshing Wang, Haoxi Chai, Yingli Zhong, Yun Shen, Wannian Yang, Junping Chen, Zhanguo Xin, Huazhong Shi, Ian Dodd
Journal of Experimental Botany.2020; 71(4): 1598. CrossRef - Unraveling the RNA Binding Properties of the Iron–Sulfur Zinc Finger Protein CPSF30
Jordan D. Pritts, Matthew S. Hursey, Jamie L. Michalek, Sharon Batelu, Timothy L. Stemmler, Sarah L. J. Michel
Biochemistry.2020; 59(8): 970. CrossRef - Stiff matrix instigates type I collagen biogenesis by mammalian cleavage factor I complex-mediated alternative polyadenylation
Zijing Zhou, Jing Qu, Li He, Yi Zhu, Shan-Zhong Yang, Feng Zhang, Ting Guo, Hong Peng, Ping Chen, Yong Zhou
JCI Insight.2020;[Epub] CrossRef - PolyA-miner: accurate assessment of differential alternative poly-adenylation from 3′Seq data using vector projections and non-negative matrix factorization
Hari Krishna Yalamanchili, Callison E Alcott, Ping Ji, Eric J Wagner, Huda Y Zoghbi, Zhandong Liu
Nucleic Acids Research.2020; 48(12): e69. CrossRef - SNP2APA: a database for evaluating effects of genetic variants on alternative polyadenylation in human cancers
Yanbo Yang, Qiong Zhang, Ya-Ru Miao, Jiajun Yang, Wenqian Yang, Fangda Yu, Dongyang Wang, An-Yuan Guo, Jing Gong
Nucleic Acids Research.2020; 48(D1): D226. CrossRef - Template-switching artifacts resemble alternative polyadenylation
Zsolt Balázs, Dóra Tombácz, Zsolt Csabai, Norbert Moldován, Michael Snyder, Zsolt Boldogkői
BMC Genomics.2019;[Epub] CrossRef - Tissue‐specific mechanisms of alternative polyadenylation: Testis, brain, and beyond (2018 update)
Clinton C. MacDonald
WIREs RNA.2019;[Epub] CrossRef - Alternative cleavage and polyadenylation in health and disease
Andreas J. Gruber, Mihaela Zavolan
Nature Reviews Genetics.2019; 20(10): 599. CrossRef - The Length of the Expressed 3′ UTR Is an Intermediate Molecular Phenotype Linking Genetic Variants to Complex Diseases
Elisa Mariella, Federico Marotta, Elena Grassi, Stefano Gilotto, Paolo Provero
Frontiers in Genetics.2019;[Epub] CrossRef
- PABPN1 loss-of-function causes APA-shift in oculopharyngeal muscular dystrophy

- A Novel Cytosolic Isoform of Mitochondrial Trans-2-Enoyl-CoA Reductase Enhances Peroxisome Proliferator-Activated Receptor α Activity
- Dong-Gyu Kim, Jae Cheal Yoo, Eunju Kim, Young-Sun Lee, Oleg V. Yarishkin, Da Yong Lee, Kun Ho Lee, Seong-Geun Hong, Eun Mi Hwang, Jae-Yong Park
- Endocrinol Metab. 2014;29(2):185-194. Published online June 26, 2014
- DOI: https://doi.org/10.3803/EnM.2014.29.2.185
- 4,254 View
- 45 Download
- 20 Web of Science
- 21 Crossref
-
 Abstract
Abstract
 PDF
PDF PubReader
PubReader Background Mitochondrial trans-2-enoyl-CoA reductase (MECR) is involved in mitochondrial synthesis of fatty acids and is highly expressed in mitochondria. MECR is also known as nuclear receptor binding factor-1, which was originally reported with yeast two-hybrid screening as a binding protein of the nuclear hormone receptor peroxisome proliferator-activated receptor α (PPARα). However, MECR and PPARα are localized at different compartment, mitochondria, and the nucleus, respectively. Therefore, the presence of a cytosolic or nuclear isoform of MECR is necessary for functional interaction between MECR and PPARα.
Methods To identify the expression pattern of MECR and the cytosolic form of MECR (cMECR), we performed reverse transcription polymerase chain reaction (RT-PCR) with various tissue samples from Sprague-Dawley rats. To confirm the interaction between cMECR and PPARα, we performed several binding assays such as yeast two-hybrid, coimmunoprecipitation, and bimolecular fluorescence complementation. To observe subcellular localization of these proteins, immunocytochemistry was performed. A luciferase assay was used to measure PPARα activity.
Results We provide evidence of an alternatively spliced variant of the rat MECR gene that yields cMECR. The cMECR lacks the N-terminal 76 amino acids of MECR and shows uniform distribution in the cytoplasm and nucleus of HeLa cells. cMECR directly bound PPARα in the nucleus and increased PPARα-dependent luciferase activity in HeLa cells.
Conclusion We found the cytosolic form of MECR (cMECR) was expressed in the cytosolic and/or nuclear region, directly binds with PPARα, and enhances PPARα activity.
-
Citations
Citations to this article as recorded by- Metabolism of phenolics in coffee and plant-based foods by canonical pathways: an assessment of the role of fatty acid β-oxidation to generate biologically-active and -inactive intermediates
Michael N. Clifford, Laurence J. King, Asimina Kerimi, Maria Gema Pereira-Caro, Gary Williamson
Critical Reviews in Food Science and Nutrition.2024; 64(11): 3326. CrossRef - Comparison of muscle nutritional composition, texture quality, carotenoid metabolites and transcriptome to underling muscle quality difference between wild-caught and pond-cultured Yellow River carp (Cyprinus carpio haematopterus)
Luming Wang, Jinrui Xiong, Chunchu Xu, Chaobin Qin, Yuru Zhang, Liping Yang, Shaoyang Zhi, Jianxin Feng, Guoxing Nie
Aquaculture.2024; 581: 740392. CrossRef - Effects of microcystin-LR on immune function, lipid metabolism and intestinal microbial structure in Eriocheir sinensis
Jinliang Du, Liping Cao, Jiancao Gao, Zhijuan Nie, Quanjie Li, Yi Sun, Nailin Shao, Jiawen Hu, Lin Zhou, Guojun Yin, Gangchun Xu
Aquaculture Reports.2024; 35: 101994. CrossRef - A defect in mitochondrial fatty acid synthesis impairs iron metabolism and causes elevated ceramide levels
Debdeep Dutta, Oguz Kanca, Seul Kee Byeon, Paul C. Marcogliese, Zhongyuan Zuo, Rishi V. Shridharan, Jun Hyoung Park, Guang Lin, Ming Ge, Gali Heimer, Jennefer N. Kohler, Matthew T. Wheeler, Benny A. Kaipparettu, Akhilesh Pandey, Hugo J. Bellen
Nature Metabolism.2023; 5(9): 1595. CrossRef - Alternative splicing liberates a cryptic cytoplasmic isoform of mitochondrial MECR that antagonizes influenza virus
Steven F. Baker, Helene Meistermann, Manuel Tzouros, Aaron Baker, Sabrina Golling, Juliane Siebourg Polster, Mitchell P. Ledwith, Anthony Gitter, Angelique Augustin, Hassan Javanbakht, Andrew Mehle, Frank Kirchhoff
PLOS Biology.2022; 20(12): e3001934. CrossRef - Genetic variants in ALDH1L1 and GLDC influence the serine-to-glycine ratio in Hispanic children
Sergey A Krupenko, Shelley A Cole, Ruixue Hou, Karin Haack, Sandra Laston, Nitesh R Mehta, Anthony G Comuzzie, Nancy F Butte, V Saroja Voruganti
The American Journal of Clinical Nutrition.2022; 116(2): 500. CrossRef - Simultaneous Presentation of Multiple Myeloma and Lung Cancer: Case Report and Gene Bioinformatics Analysis
Ping-Ping Xiao, Bing-Qing Luo, Wei Fan, Xu-Yan Chen, Zhi-Gao Dong, Jin-Mei Huang, Yi Zhang, Yong-Quan Chen
Frontiers in Oncology.2022;[Epub] CrossRef - Fatty acid metabolism-related genes are associated with flavor-presenting aldehydes in Chinese local chicken
Xiaoya Yuan, Huanxian Cui, Yuxi Jin, Wenjuan Zhao, Xiaojing Liu, Yongli Wang, Jiqiang Ding, Li Liu, Jie Wen, Guiping Zhao
Frontiers in Genetics.2022;[Epub] CrossRef - NRBF2-mediated autophagy contributes to metabolite replenishment and radioresistance in glioblastoma
Jeongha Kim, Hyunkoo Kang, Beomseok Son, Min-Jung Kim, JiHoon Kang, Kang Hyun Park, Jaewan Jeon, Sunmi Jo, Hae Yu Kim, HyeSook Youn, BuHyun Youn
Experimental & Molecular Medicine.2022; 54(11): 1872. CrossRef - Mitochondrial Fatty Acids and Neurodegenerative Disorders
Alexander J. Kastaniotis, Kaija J. Autio, Remya R. Nair
The Neuroscientist.2021; 27(2): 143. CrossRef - The effects of chronic cadmium exposure on Bufo gargarizans larvae: Histopathological impairment, gene expression alteration and fatty acid metabolism disorder in the liver
Zongqi Ju, Jing Ya, Xinyi Li, Hongyuan Wang, Hongfeng Zhao
Aquatic Toxicology.2020; 222: 105470. CrossRef - Exploration of targets regulated by miR-125b in porcine adipocytes
Xiao Cheng, Xingping Chen, Peng Wang, Ting Chen, Jiajie Sun, Qianyun Xi, Yongliang Zhang
In Vitro Cellular & Developmental Biology - Animal.2020; 56(2): 103. CrossRef - Mitochondrial fatty acid synthesis coordinates oxidative metabolism in mammalian mitochondria
Sara M Nowinski, Ashley Solmonson, Scott F Rusin, J Alan Maschek, Claire L Bensard, Sarah Fogarty, Mi-Young Jeong, Sandra Lettlova, Jordan A Berg, Jeffrey T Morgan, Yeyun Ouyang, Bradley C Naylor, Joao A Paulo, Katsuhiko Funai, James E Cox, Steven P Gygi,
eLife.2020;[Epub] CrossRef - Polymorphisms in ten candidate genes are associated with conformational and locomotive traits in Spanish Purebred horses
Natalia Sevane, Susana Dunner, Ana Boado, Javier Cañon
Journal of Applied Genetics.2017; 58(3): 355. CrossRef - Deep RNA sequencing of pectoralis muscle transcriptomes during late-term embryonic to neonatal development in indigenous Chinese duck breeds
Chunhong Zhu, Weitao Song, Zhiyun Tao, Hongxiang Liu, Wenjuan Xu, Shuangjie Zhang, Huifang Li, Cristina Óvilo
PLOS ONE.2017; 12(8): e0180403. CrossRef - Mitochondrial fatty acid synthesis, fatty acids and mitochondrial physiology
Alexander J. Kastaniotis, Kaija J. Autio, Juha M. Kerätär, Geoffray Monteuuis, Anne M. Mäkelä, Remya R. Nair, Laura P. Pietikäinen, Antonina Shvetsova, Zhijun Chen, J. Kalervo Hiltunen
Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids.2017; 1862(1): 39. CrossRef - Genetic modifications of Mecr reveal a role for mitochondrial 2-enoyl-CoA/ACP reductase in placental development in mice
Remya R. Nair, Juha M. Kerätär, Kaija J. Autio, Ali J. Masud, Mikko A.J. Finnilä, Helena I. Autio-Harmainen, Ilkka J. Miinalainen, Pentti A. Nieminen, J. Kalervo Hiltunen, Alexander J. Kastaniotis
Human Molecular Genetics.2017; 26(11): 2104. CrossRef - MECR Mutations Cause Childhood-Onset Dystonia and Optic Atrophy, a Mitochondrial Fatty Acid Synthesis Disorder
Gali Heimer, Juha M. Kerätär, Lisa G. Riley, Shanti Balasubramaniam, Eran Eyal, Laura P. Pietikäinen, J. Kalervo Hiltunen, Dina Marek-Yagel, Jeffrey Hamada, Allison Gregory, Caleb Rogers, Penelope Hogarth, Martha A. Nance, Nechama Shalva, Alvit Veber, Mic
The American Journal of Human Genetics.2016; 99(6): 1229. CrossRef - Genome‐wide association study with the risk of schizophrenia in a Korean population
Lyoung Hyo Kim, Byung Lae Park, Hyun Sub Cheong, Suhg Namgoong, Ji On Kim, Jeong‐Hyun Kim, Joong‐Gon Shin, Chul Soo Park, Bong‐Jo Kim, Jae Won Kim, Ihn‐Geun Choi, Jaeuk Hwang, Hyoung Doo Shin, Sung‐Il Woo
American Journal of Medical Genetics Part B: Neuropsychiatric Genetics.2016; 171(2): 257. CrossRef - A global transcriptional analysis of Megalobrama amblycephala revealing the molecular determinants of diet-induced hepatic steatosis
Dingdong Zhang, Kangle Lu, Guangzhen Jiang, Wenbin Liu, Zaijie Dong, Hongyan Tian, Xiangfei Li
Gene.2015; 570(2): 255. CrossRef - Articles in 'Endocrinology and Metabolism' in 2014
Won-Young Lee
Endocrinology and Metabolism.2015; 30(1): 47. CrossRef
- Metabolism of phenolics in coffee and plant-based foods by canonical pathways: an assessment of the role of fatty acid β-oxidation to generate biologically-active and -inactive intermediates

- Obesity and Metabolism
- Regulation of Adipocyte Differentiation via MicroRNAs
- You Hwa Son, Sojeong Ka, A Young Kim, Jae Bum Kim
- Endocrinol Metab. 2014;29(2):122-135. Published online June 26, 2014
- DOI: https://doi.org/10.3803/EnM.2014.29.2.122
- 6,752 View
- 104 Download
- 74 Web of Science
- 72 Crossref
-
 Abstract
Abstract
 PDF
PDF PubReader
PubReader Adipocyte differentiation, termed adipogenesis, is a complicated process in which pluripotent mesenchymal stem cells differentiate into mature adipocytes. The process of adipocyte differentiation is tightly regulated by a number of transcription factors, hormones and signaling pathway molecules. Recent studies have demonstrated that microRNAs, which belong to small noncoding RNA species, are also involved in adipocyte differentiation.
In vivo andin vitro studies have revealed that various microRNAs affect adipogenesis by targeting several adipogenic transcription factors and key signaling molecules. In this review, we will summarize the roles of microRNAs in adipogenesis and their target genes associated with each stage of adipocyte differentiation.-
Citations
Citations to this article as recorded by- A review of the role of transcription factors in regulating adipogenesis and lipogenesis in beef cattle
Belete Kuraz Abebe, Hongbao Wang, Anning Li, Linsen Zan
Journal of Animal Breeding and Genetics.2024; 141(3): 235. CrossRef - A review of the role of epigenetic studies for intramuscular fat deposition in beef cattle
Belete Kuraz Abebe, Jianfang Wang, Juntao Guo, Hongbao Wang, Anning Li, Linsen Zan
Gene.2024; 908: 148295. CrossRef - Transcriptome analysis of miRNAs during myoblasts adipogenic differentiation
Chengchuang Song, Xue Fang, Qi Wang, Yaqi Chen, Bei Zhao, Yanhong Wang, Xingtang Fang, Chunlei Zhang
Animal Biotechnology.2023; 34(4): 1406. CrossRef - Advances in the regulation of adipogenesis and lipid metabolism by exosomal ncRNAs and their role in related metabolic diseases
Cong Liu, Xilin Liu, Hong Li, Zhichen Kang
Frontiers in Cell and Developmental Biology.2023;[Epub] CrossRef - Indian gooseberry and barley sprout mixture prevents obesity by regulating adipogenesis, lipogenesis, and lipolysis in C57BL/6J mice with high-fat diet-induced obesity
Soo-Jeung Park, Jong-Lae Kim, Mi-Ryeong Park, Jong Wook Lee, Ok-Kyung Kim, Jeongmin Lee
Journal of Functional Foods.2022; 90: 104951. CrossRef - Interleukin-6 mimics insulin-dependent cellular distribution of some cytoskeletal proteins and Glut4 transporter without effect on glucose uptake in 3T3-L1 adipocytes
Maciej Błaszczyk, Małgorzata Gajewska, Marta Dymowska, Alicja Majewska, Tomasz Domoradzki, Adam Prostek, Rafał Pingwara, Magdalena Hulanicka, Katarzyna Grzelkowska-Kowalczyk
Histochemistry and Cell Biology.2022;[Epub] CrossRef - Heat-Killed Enterococcus faecalis Prevents Adipogenesis and High Fat Diet-Induced Obesity by Inhibition of Lipid Accumulation through Inhibiting C/EBP-α and PPAR-γ in the Insulin Signaling Pathway
Jin-Ho Lee, Keun-Jung Woo, Min-Ah Kim, Joonpyo Hong, Jihee Kim, Sun-Hong Kim, Kwon-Il Han, Masahiro Iwasa, Tack-Joong Kim
Nutrients.2022; 14(6): 1308. CrossRef - The delivery of miR-21a-5p by extracellular vesicles induces microglial polarization via the STAT3 pathway following hypoxia-ischemia in neonatal mice
Dan-Qing Xin, Yi-Jing Zhao, Ting-Ting Li, Hong-Fei Ke, Cheng-Cheng Gai, Xiao-Fan Guo, Wen-Qiang Chen, De-Xiang Liu, Zhen Wang
Neural Regeneration Research.2022; 17(10): 2238. CrossRef - Extracellular Vesicles from Adipose Tissue Could Promote Metabolic Adaptation through PI3K/Akt/mTOR
Jaime Delgadillo-Velázquez, Herminia Mendivil-Alvarado, Carlos Daniel Coronado-Alvarado, Humberto Astiazaran-Garcia
Cells.2022; 11(11): 1831. CrossRef - Adipocyte differentiation between obese and lean conditions depends on changes in miRNA expression
Yerim Heo, Hyunjung Kim, Jiwon Lim, Sun Shim Choi
Scientific Reports.2022;[Epub] CrossRef - Comparison of Selected Non-Coding RNAs and Gene Expression Profiles between Common Osteosarcoma Cell Lines
Mateusz Sikora, Katarzyna Krajewska, Klaudia Marcinkowska, Anna Raciborska, Rafał Jakub Wiglusz, Agnieszka Śmieszek
Cancers.2022; 14(18): 4533. CrossRef - Mesenchymal Stem Cell Secreted-Extracellular Vesicles are Involved in Chondrocyte Production and Reduce Adipogenesis during Stem Cell Differentiation
Yu-Chen Tsai, Tai-Shan Cheng, Hsiu-Jung Liao, Ming-Hsi Chuang, Hui-Ting Chen, Chun-Hung Chen, Kai-Ling Zhang, Chih-Hung Chang, Po-Cheng Lin, Chi-Ying F. Huang
Tissue Engineering and Regenerative Medicine.2022; 19(6): 1295. CrossRef - Tissue and circulating microRNAs as biomarkers of response to obesity treatment strategies
G. Catanzaro, T. Filardi, C. Sabato, A. Vacca, S. Migliaccio, S. Morano, E. Ferretti
Journal of Endocrinological Investigation.2021; 44(6): 1159. CrossRef - Benzyl Butyl Phthalate Induced Early lncRNA H19 Regulation in C3H10T1/2 Stem Cell Line
Jian Zhang, Mahua Choudhury
Chemical Research in Toxicology.2021; 34(1): 54. CrossRef - Inhibition of preadipocyte differentiation by Lycium barbarum polysaccharide treatment in 3T3-L1 cultures
Xiaochun Xu, Wenjuan Chen, Shukun Yu, Qian Lei, Lihong Han, Wenping Ma
Electronic Journal of Biotechnology.2021; 50: 53. CrossRef - MiR-25-3p regulates the differentiation of intramuscular preadipocytes in goat via targeting <i>KLF4</i>
Yu Du, Yue Zhao, Yong Wang, Qingyong Meng, Jiangjiang Zhu, Yaqiu Lin
Archives Animal Breeding.2021; 64(1): 17. CrossRef - MiR-208b Regulates Rabbit Preadipocyte Proliferation and Differentiation
Jiahao Shao, Ting Pan, Jie Wang, Tao Tang, Yanhong Li, Xianbo Jia, Songjia Lai
Genes.2021; 12(6): 890. CrossRef - Comparing the effect of cinnamaldehyde and metformin on expression of MiR320 and MiR26-b in insulin resistant 3T3L1 adipocytes
Yousof Naghiaee, Mahmood Vakili, Mohammad Mohammadi, Azra Mohiti, Javad Mohiti-Ardakani
Phytomedicine Plus.2021; 1(4): 100122. CrossRef - Lower miR‐26a levels in breastmilk affect gene expression in adipose tissue of offspring
Catalina A. Pomar, Francisca Serra, Andreu Palou, Juana Sánchez
The FASEB Journal.2021;[Epub] CrossRef - MicroRNA-378 regulates adipogenic differentiation in bovine intramuscular preadipocytes by targeting CaMKK2
Dongwei Li, Heng Wang, Yongmin Li, Changqing Qu, Yunhai Zhang, Hongyu Liu, Xiaorong Zhang
Adipocyte.2021; 10(1): 483. CrossRef - miR-6315 Attenuates Methotrexate Treatment-Induced Decreased Osteogenesis and Increased Adipogenesis Potentially through Modulating TGF-β/Smad2 Signalling
Ya-Li Zhang, Liang Liu, Yu-Wen Su, Cory J. Xian
Biomedicines.2021; 9(12): 1926. CrossRef - Inhibitory Effects of Pinostilbene on Adipogenesis in 3T3-L1 Adipocytes: A Study of Possible Mechanisms
You Chul Chung, Chang-Gu Hyun
International Journal of Molecular Sciences.2021; 22(24): 13446. CrossRef - Regulation of Methylase METTL3 on Fat Deposition
Gang Luo, Jialing Chen, Zhanjun Ren
Diabetes, Metabolic Syndrome and Obesity: Targets and Therapy.2021; Volume 14: 4843. CrossRef - miR-214-5p Regulating Differentiation of Intramuscular Preadipocytes in Goats via Targeting KLF12
Yu Du, Yong Wang, Yanyan Li, Quzhe Emu, Jiangjiang Zhu, Yaqiu Lin
Frontiers in Genetics.2021;[Epub] CrossRef - Screening and identification of MicroRNAs expressed in perirenal adipose tissue during rabbit growth
Guoze Wang, Guo Guo, Xueting Tian, Shenqiang Hu, Kun Du, Qinghai Zhang, Jingxin Mao, Xianbo Jia, Shiyi Chen, Jie Wang, Songjia Lai
Lipids in Health and Disease.2020;[Epub] CrossRef - Descending Expression of miR320 in Insulin-Resistant Adipocytes Treated with Ascending Concentrations of Metformin
Yousof Naghiaee, Reza Didehdar, Zahra Malekpour-Dehkordi, Fatemeh Pourrajab, Javad Mohiti-Ardakani
Biochemical Genetics.2020; 58(5): 661. CrossRef - Citrus aurantium L. Dry Extracts Ameliorate Adipocyte Differentiation of 3T3-L1 Cells Exposed to TNFα by Down-Regulating miR-155 Expression
Michele Campitelli, Antonella Desiderio, Giuseppe Cacace, Cecilia Nigro, Immacolata Prevenzano, Alessia Leone, Sonia de Simone, Pietro Campiglia, Pietro Formisano, Gregory A. Raciti, Francesco Beguinot, Claudia Miele
Nutrients.2020; 12(6): 1587. CrossRef - Metabolic Benefits of MicroRNA-22 Inhibition
Marc Thibonnier, Christine Esau
Nucleic Acid Therapeutics.2020; 30(2): 104. CrossRef - Differentially Expressed miRNA-Gene Targets Related to Intramuscular Fat in Musculus Longissimus Dorsi of Charolais × Holstein F2-Crossbred Bulls
Bilal Ahmad Mir, Henry Reyer, Katrin Komolka, Siriluck Ponsuksili, Christa Kühn, Steffen Maak
Genes.2020; 11(6): 700. CrossRef - MiR224-5p Inhibitor Restrains Neuronal Apoptosis by Targeting NR4A1 in the Oxygen-Glucose Deprivation (OGD) Model
Ling-Ling Liu, Shan Qiao, Mei-Ling Wang, Huai-Kuan Wu, Yong-Xin Su, Ke-Mo Wang, Xue-Wu Liu
Frontiers in Neuroscience.2020;[Epub] CrossRef - Adipogenesis: A Complex Interplay of Multiple Molecular Determinants and Pathways
Melvin A. Ambele, Priyanka Dhanraj, Rachel Giles, Michael S. Pepper
International Journal of Molecular Sciences.2020; 21(12): 4283. CrossRef - MicroRNA‑449a regulates the progression of brain aging by targeting SCN2B in SAMP8 mice
Ya‑Xin Tan, Ying Hong, Shui Jiang, Min‑Nan Lu, Shan Li, Bo Chen, Li Zhang, Tao Hu, Rui Mao, Rong Mei, Yan‑Bin Xiyang
International Journal of Molecular Medicine.2020;[Epub] CrossRef - Influence of adenovirus 36 seropositivity on the expression of adipogenic microRNAs in obese subjects
Víctor Manríquez, Alvaro Gutierrez, Alexis Morales, Roberto Brito, Monica Pavez, Jorge Sapunar, Luis Fonseca, Víctor Molina, Eugenia Ortiz, Maria Ines Barra, Camila Reimer, Maria Charles, Constance Schneider, Alvaro Cerda
International Journal of Obesity.2020; 44(11): 2303. CrossRef - Metformin downregulates miR223 expression in insulin-resistant 3T3L1 cells and human diabetic adipose tissue
Yousof Naghiaee, Reza Didehdar, Fatemeh Pourrajab, Masoud Rahmanian, Naeime Heiranizadeh, Azra Mohiti, Javad Mohiti-Ardakani
Endocrine.2020; 70(3): 498. CrossRef - Metabolic and energetic benefits of microRNA-22 inhibition
Marc Thibonnier, Christine Esau, Sujoy Ghosh, Edward Wargent, Claire Stocker
BMJ Open Diabetes Research & Care.2020; 8(1): e001478. CrossRef - Bta-miR-376a Targeting KLF15 Interferes with Adipogenesis Signaling Pathway to Promote Differentiation of Qinchuan Beef Cattle Preadipocytes
Xingyi Chen, Sayed Haidar Abbas Raza, Gong Cheng, Xinhao Ma, Jianfang Wang, Linsen Zan
Animals.2020; 10(12): 2362. CrossRef - The cross-talk between adipokines and miRNAs in health and obesity-mediated diseases
Ahmad Ghasemi, Seyed Isaac Hashemy, Mohsen Azimi-Nezhad, Alireza Dehghani, Jafar Saeidi, Mahnaz Mohtashami
Clinica Chimica Acta.2019; 499: 41. CrossRef - MicroRNAs and long noncoding RNAs: new regulators in cell fate determination of mesenchymal stem cells
Zixiang Wu, Shujing Liang, Wenyu Kuai, Lifang Hu, Airong Qian
RSC Advances.2019; 9(64): 37300. CrossRef - Harnessing adipogenesis to prevent obesity
Nida Haider, Louise Larose
Adipocyte.2019; 8(1): 98. CrossRef - MiR‐127 attenuates adipogenesis by targeting MAPK4 and HOXC6 in porcine adipocytes
Yun Gao, Yingqian Wang, Xiaochang Chen, Ying Peng, Fenfen Chen, Yulin He, Weijun Pang, Gongshe Yang, Taiyong Yu
Journal of Cellular Physiology.2019; 234(12): 21838. CrossRef - MicroRNA-425 controls lipogenesis and lipolysis in adipocytes
Renli Qi, Jing Wang, Qi Wang, Xiaoyu Qiu, Feiyun Yang, Zuohua Liu, Jinxiu Huang
Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids.2019; 1864(5): 744. CrossRef - miRNA-7a-2-3p Inhibits Neuronal Apoptosis in Oxygen-Glucose Deprivation (OGD) Model
Zi-Bin Zhang, Ya-Xin Tan, Qiong Zhao, Liu-Lin Xiong, Jia Liu, Fei-Fei Xu, Yang Xu, Larisa Bobrovskaya, Xin-Fu Zhou, Ting-Hua Wang
Frontiers in Neuroscience.2019;[Epub] CrossRef - IL‐1α inhibits proliferation and adipogenic differentiation of human adipose‐derived mesenchymal stem cells through NF‐κB‐ and ERK1/2‐mediated proinflammatory cytokines
Xuerong Sun, Tangbin Zou, Changqing Zuo, Mingmeng Zhang, Benyan Shi, Zhiwen Jiang, Hongjing Cui, Xiaoxin Liao, Xiaoyi Li, Yuelian Tang, Yusheng Liu, Xinguang Liu
Cell Biology International.2018; 42(7): 794. CrossRef - MicroRNA‐224‐5p regulates adipocyte apoptosis induced by TNFα via controlling NF‐κB activation
Renli Qi, Jinxiu Huang, Qi Wang, Hong Liu, Ruisheng Wang, Jing Wang, Feiyun Yang
Journal of Cellular Physiology.2018; 233(2): 1236. CrossRef - Potential role of microRNAs in the regulation of adipocytes liposecretion and adipose tissue physiology
Giulia Maurizi, Lucia Babini, Lucio Della Guardia
Journal of Cellular Physiology.2018; 233(12): 9077. CrossRef - Chronic hyperinsulinemia induced miR-27b is linked to adipocyte insulin resistance by targeting insulin receptor
Ankita Srivastava, Kripa Shankar, Muheeb Beg, Sujith Rajan, Abhishek Gupta, Salil Varshney, Durgesh Kumar, Sanchita Gupta, Raj Kumar Mishra, Anil Nilkanth Gaikwad
Journal of Molecular Medicine.2018; 96(3-4): 315. CrossRef - Bta-miR-130a/b regulates preadipocyte differentiation by targeting PPARG and CYP2U1 in beef cattle
Xueyao Ma, Dawei Wei, Gong Cheng, Shijun Li, Li Wang, Yaning Wang, Xiaoyu Wang, Song Zhang, Hongbao Wang, Linsen Zan
Molecular and Cellular Probes.2018; 42: 10. CrossRef - Transdifferentiation of adipocytes to osteoblasts: potential for orthopaedic treatment
Daphne P L Lin, Crispin R Dass
Journal of Pharmacy and Pharmacology.2018; 70(3): 307. CrossRef - Characterization of miRNA transcriptome profiles related to breast muscle development and intramuscular fat deposition in chickens
Shouyi Fu, Yinli Zhao, Yuanfang Li, Guoxi Li, Yi Chen, Zhuanjian Li, Guirong Sun, Hong Li, Xiangtao Kang, Fengbin Yan
Journal of Cellular Biochemistry.2018; 119(8): 7063. CrossRef - Transcriptomic Analyses of Adipocyte Differentiation From Human Mesenchymal Stromal‐Cells (MSC)
Antonio Casado‐Díaz, Jaouad Anter, Sören Müller, Peter Winter, José Manuel Quesada‐Gómez, Gabriel Dorado
Journal of Cellular Physiology.2017; 232(4): 771. CrossRef - Downregulated miR-29a/b/c during Contact Inhibition Stage Promote 3T3-L1 Adipogenesis by Targeting DNMT3A
Yingjie Zhu, Guangyong Zheng, Huichao Wang, Yudong Jia, Ying Zhang, Yanfeng Tang, Wenlong Li, Yanan Fan, Xiaodong Zhang, Youwen Liu, Sanhong Liu, Makoto Kanzaki
PLOS ONE.2017; 12(1): e0170636. CrossRef - Potential miRNA involvement in the anti-adipogenic effect of resveratrol and its metabolites
Itziar Eseberri, Arrate Lasa, Jonatan Miranda, Ana Gracia, Maria P. Portillo, Cristina Óvilo
PLOS ONE.2017; 12(9): e0184875. CrossRef - Identification and characterization of differentially expressed miRNAs in subcutaneous adipose between Wagyu and Holstein cattle
Yuntao Guo, Xiuxiu Zhang, Wanlong Huang, Xiangyang Miao
Scientific Reports.2017;[Epub] CrossRef - Integrated analysis of mRNA and miRNA expression profiles in livers of Yimeng black pigs with extreme phenotypes for backfat thickness
Wentong Li, Yalan Yang, Ying Liu, Shuai Liu, Xiuxiu Li, Yingping Wang, Yanmin Zhang, Hui Tang, Rong Zhou, Kui Li
Oncotarget.2017; 8(70): 114787. CrossRef - Small non coding RNAs in adipocyte biology and obesity
Ez-Zoubir Amri, Marcel Scheideler
Molecular and Cellular Endocrinology.2017; 456: 87. CrossRef - Role of MicroRNA Regulation in Obesity-Associated Breast Cancer: Nutritional Perspectives
Ravi Kasiappan, Dheeran Rajarajan
Advances in Nutrition.2017; 8(6): 868. CrossRef - Biomolecular features of inflammation in obese rheumatoid arthritis patients: management considerations
Barbara Tolusso, Stefano Alivernini, Maria Rita Gigante, Gianfranco Ferraccioli, Elisa Gremese
Expert Review of Clinical Immunology.2016; 12(7): 751. CrossRef - Circulating microRNAs are deregulated in overweight/obese children: preliminary results of the I.Family study
Giuseppe Iacomino, Paola Russo, Ilaria Stillitano, Fabio Lauria, Pasquale Marena, Wolfgang Ahrens, Pasquale De Luca, Alfonso Siani
Genes & Nutrition.2016;[Epub] CrossRef - Role of MicroRNAs in NAFLD/NASH
Gyongyi Szabo, Timea Csak
Digestive Diseases and Sciences.2016; 61(5): 1314. CrossRef - MicroRNA‐29b promotes the adipogenic differentiation of human adipose tissue‐derived stromal cells
Xi‐Mei Zhang, Li‐Hong Wang, Dong‐Ju Su, Dan Zhu, Qiu‐Ming Li, Mei‐Hua Chi
Obesity.2016; 24(5): 1097. CrossRef - MiR-181a-5p regulates 3T3-L1 cell adipogenesis by targeting <italic>Smad7</italic> and <italic>Tcf7l2</italic>
Dan Ouyang, Lifeng Xu, Lihua Zhang, Dongguang Guo, Xiaotong Tan, Xiaofang Yu, Junjie Qi, Yaqiong Ye, Qihong Liu, Yongjiang Ma, Yugu Li
Acta Biochimica et Biophysica Sinica.2016; 48(11): 1034. CrossRef - Metformin-suppressed differentiation of human visceral preadipocytes: Involvement of microRNAs
Koji Fujita, Hisakazu Iwama, Kyoko Oura, Tomoko Tadokoro, Kayo Hirose, Miwako Watanabe, Teppei Sakamoto, Akiko Katsura, Shima Mimura, Takako Nomura, Joji Tani, Hisaaki Miyoshi, Asahiro Morishita, Hirohito Yoneyama, Keiichi Okano, Yasuyuki Suzuki, Takashi
International Journal of Molecular Medicine.2016; 38(4): 1135. CrossRef - Systematic study of cis-antisense miRNAs in animal species reveals miR-3661 to target PPP2CA in human cells
Jian Wang, Zongcheng Li, Bailong Liu, Guangnan Chen, Ningsheng Shao, Xiaomin Ying, Ya Wang
RNA.2016; 22(1): 87. CrossRef - Oxidative stress, redox regulation and diseases of cellular differentiation
Zhi-Wei Ye, Jie Zhang, Danyelle M. Townsend, Kenneth D. Tew
Biochimica et Biophysica Acta (BBA) - General Subjects.2015; 1850(8): 1607. CrossRef - Noncoding RNAs, cytokines, and inflammation-related diseases
José Luiz Marques-Rocha, Mirian Samblas, Fermin I. Milagro, Josefina Bressan, J. Alfredo Martínez, Amelia Marti
The FASEB Journal.2015; 29(9): 3595. CrossRef - Polymorphism in miR-31 and miR-584 binding site in the angiotensinogen gene differentially influences body fat distribution in both sexes
Jan Machal, Jan Novak, Renata Hezova, Filip Zlamal, Anna Vasku, Ondrej Slaby, Julie Bienertova-Vasku
Genes & Nutrition.2015;[Epub] CrossRef - Mitochondria-related miR-141-3p contributes to mitochondrial dysfunction in HFD-induced obesity by inhibiting PTEN
Juan Ji, Yufeng Qin, Jing Ren, Chuncheng Lu, Rong Wang, Xiuliang Dai, Ran Zhou, Zhenyao Huang, Miaofei Xu, Minjian Chen, Wei Wu, Ling Song, Hongbing Shen, Zhibin Hu, Dengshun Miao, Yankai Xia, Xinru Wang
Scientific Reports.2015;[Epub] CrossRef - MicroRNAs in Bone Balance and Osteoporosis
Junying Chen, Min Qiu, Ce Dou, Zhen Cao, Shiwu Dong
Drug Development Research.2015; 76(5): 235. CrossRef - Articles in 'Endocrinology and Metabolism' in 2014
Won-Young Lee
Endocrinology and Metabolism.2015; 30(1): 47. CrossRef - MicroRNA regulatory networks in human adipose tissue and obesity
Peter Arner, Agné Kulyté
Nature Reviews Endocrinology.2015; 11(5): 276. CrossRef - Epigenomics, gestational programming and risk of metabolic syndrome
M Desai, J K Jellyman, M G Ross
International Journal of Obesity.2015; 39(4): 633. CrossRef - Expression Profiling and Structural Characterization of MicroRNAs in Adipose Tissues of Hibernating Ground Squirrels
Cheng-Wei Wu, Kyle K. Biggar, Kenneth B. Storey
Genomics, Proteomics & Bioinformatics.2014; 12(6): 284. CrossRef
- A review of the role of transcription factors in regulating adipogenesis and lipogenesis in beef cattle

- Adrenal gland
- The Molecular Pathogenesis of Pituitary Adenomas: An Update
- Xiaobing Jiang, Xun Zhang
- Endocrinol Metab. 2013;28(4):245-254. Published online December 12, 2013
- DOI: https://doi.org/10.3803/EnM.2013.28.4.245
- 4,076 View
- 43 Download
- 37 Crossref
-
 Abstract
Abstract
 PDF
PDF PubReader
PubReader Pituitary tumors represent the most common intracranial neoplasms accompanying serious morbidity through mass effects and inappropriate secretion of pituitary hormones. Understanding the etiology of pituitary tumorigenesis will facilitate the development of satisfactory treatment for pituitary adenomas. Although the pathogenesis of pituitary adenomas is largely unknown, considerable evidence indicates that the pituitary tumorigenesis is a complex process involving multiple factors, including genetic and epigenetic changes. This review summarized the recent progress in the study of pituitary tumorigenesis, focusing on the role of tumor suppressor genes, oncogenes and microRNAs.
-
Citations
Citations to this article as recorded by- Plurihormonal Pituitary Neuroendocrine Tumors: Clinical Relevance of Immunohistochemical Analysis
Roxana-Ioana Dumitriu-Stan, Iulia-Florentina Burcea, Ramona Dobre, Valeria Nicoleta Nastase, Raluca Amalia Ceausu, Marius Raica, Catalina Poiana
Diagnostics.2024; 14(2): 170. CrossRef - LncRNA MYMLR promotes pituitary adenoma development by upregulating carbonyl reductase 1 via sponging miR-197-3p
Tuo Wang, Ping Mao, Yan Zhang, Bo Cui, Mao-De Wang, Ya Li, Ke Gao
Anti-Cancer Drugs.2022; 33(10): 1058. CrossRef - Transcriptomic Profiles of Normal Pituitary Cells and Pituitary Neuroendocrine Tumor Cells
Jun Y. Oh, Robert C. Osorio, Jangham Jung, Luis Carrete, Nikita Choudhary, Meeki Lad, Atul Saha, Manish K. Aghi
Cancers.2022; 15(1): 110. CrossRef - EMT-Related Markers in Serum Exosomes are Potential Diagnostic Biomarkers for Invasive Pituitary Adenomas
Kelin Chen, Guoge Li, Xixiong Kang, Pinan Liu, Lingye Qian, Yijun Shi, Rasha Alsamani Osman, Zhijun Yang, Guojun Zhang
Neuropsychiatric Disease and Treatment.2021; Volume 17: 3769. CrossRef - The Role of Long Noncoding RNAs in the Biology of Pituitary Adenomas
Ozal Beylerli, Ilgiz Gareev, Valentin Pavlov, Xin Chen, Shiguang Zhao
World Neurosurgery.2020; 137: 252. CrossRef - Sellar Tumors
Katherine E. Schwetye, Sonika M. Dahiya
Surgical Pathology Clinics.2020; 13(2): 305. CrossRef - Genomic and molecular characterization of pituitary adenoma pathogenesis: review and translational opportunities
Mazin Elsarrag, Parantap D. Patel, Ajay Chatrath, Davis Taylor, John A. Jane
Neurosurgical Focus.2020; 48(6): E11. CrossRef - Metabolic profiling reveals distinct metabolic alterations in different subtypes of pituitary adenomas and confers therapeutic targets
Jie Feng, Hua Gao, Qi Zhang, Yang Zhou, Chuzhong Li, Sida Zhao, Lichuan Hong, Jinjin Yang, Shuyu Hao, Wan Hong, Zhengping Zhuang, Guowang Xu, Yazhuo Zhang
Journal of Translational Medicine.2019;[Epub] CrossRef - Quantitative Analysis of Proteome in Non-functional Pituitary Adenomas: Clinical Relevance and Potential Benefits for the Patients
Tingting Cheng, Ya Wang, Miaolong Lu, Xiaohan Zhan, Tian Zhou, Biao Li, Xianquan Zhan
Frontiers in Endocrinology.2019;[Epub] CrossRef - circOMA1-Mediated miR-145-5p Suppresses Tumor Growth of Nonfunctioning Pituitary Adenomas by Targeting TPT1
Qiu Du, Bin Hu, Yajuan Feng, Zongming Wang, Xin Wang, Dimin Zhu, Yonghong Zhu, Xiaobing Jiang, Haijun Wang
The Journal of Clinical Endocrinology & Metabolism.2019; 104(6): 2419. CrossRef - Differential Expression of HMGA1 and HMGA2 in pituitary neuroendocrine tumors
Sérgio Portovedo, Nadja Gaido, Bruno de Almeida Nunes, Ana Giselia Nascimento, Allysson Rocha, Marcelo Magalhães, Gilvan Cortes Nascimento, Denise Pires de Carvalho, Paula Soares, Christina Takiya, Manuel dos Santos Faria, Leandro Miranda-Alves
Molecular and Cellular Endocrinology.2019; 490: 80. CrossRef - Double Pituitary Adenomas with Synchronous Somatotroph and Corticotroph Clinical Presentation of Acromegaly and Cushing's Disease
Naomi Collazo-Gutiérrez, Orlando de Jesús, Maria Villamil-Jarauta, Milliette Alvarado, Loida González, Margarita Ramírez, Victor J. Carlo-Chevere
World Neurosurgery.2019; 132: 161. CrossRef - Association of ApoE haplotype with clinical evidence of pituitary adenoma
Agne Sidaraite, Alvita Vilkeviciute, Brigita Glebauskiene, Loresa Kriauciuniene, Dalia Zaliuniene, Rasa Liutkeviciene
Gene.2019; 706: 154. CrossRef - Next-generation sequencing of microRNAs reveals a unique expression pattern in different types of pituitary adenomas
Zongze He, Longyi Chen, Xiao Hu, Jian Tang, Linfu He, Junting Hu, Fan Fei, Qi Wang
Endocrine Journal.2019; 66(8): 709. CrossRef - Growth hormone and prolactin-staining tumors causing acromegaly: a retrospective review of clinical presentations and surgical outcomes
Jonathan Rick, Arman Jahangiri, Patrick M. Flanigan, Ankush Chandra, Sandeep Kunwar, Lewis Blevins, Manish K. Aghi
Journal of Neurosurgery.2019; 131(1): 147. CrossRef - Study of major genetic factors involved in pituitary tumorigenesis and their impact on clinical and biological characteristics of sporadic somatotropinomas and non-functioning pituitary adenomas
R.K. Foltran, P.V.G.H. Amorim, F.H. Duarte, I.P.P. Grande, A.C.T.B. Freire, F.P. Frassetto, J.B. Dettoni, V.A. Alves, I. Castro, E.B. Trarbach, M.D. Bronstein, R.S. Jallad
Brazilian Journal of Medical and Biological Research.2018;[Epub] CrossRef - Detection of circulating tumor cells in patients with pituitary tumors
Gao Hua, He Yanjiao, Liu Qian, Wang Jichao, Zhang Yazhuo
BMC Cancer.2018;[Epub] CrossRef - Plurihormonal ACTH-GH Pituitary Adenoma: Case Report and Systematic Literature Review
Elena Roca, Pier Paolo Mattogno, Teresa Porcelli, Luigi Poliani, Francesco Belotti, Alberto Schreiber, Filippo Maffezzoni, Marco Maria Fontanella, Francesco Doglietto
World Neurosurgery.2018; 114: e158. CrossRef - The role of galectin-3 in the tumorigenesis and progression of pituitary tumors
Bo Diao, Ying Liu, Guo‑Zheng Xu, Yi Zhang, Jun Xie, Jie Gong
Oncology Letters.2018;[Epub] CrossRef - Programmed cell senescence: role of IL-6 in the pituitary
Melanie Sapochnik, Mariana Fuertes, Eduardo Arzt
Journal of Molecular Endocrinology.2017; 58(4): R241. CrossRef - Selective molecular biomarkers to predict biologic behavior in pituitary tumors
Aydin Sav, Fabio Rotondo, Luis V. Syro, Meric A. Altinoz, Kalman Kovacs
Expert Review of Endocrinology & Metabolism.2017; 12(3): 177. CrossRef - MicroRNA-200b inhibits pituitary tumor cell proliferation and invasion by targeting PKCα
Yuanchuan Wang, Xiaohong Yin, Long Zhao, Shun Li, Jie Duan, Renzhao Kuang, Junwei Duan
Experimental and Therapeutic Medicine.2017; 14(2): 1706. CrossRef - Increased expression of the microRNA 106b~25 cluster and its host gene MCM7 in corticotroph pituitary adenomas is associated with tumor invasion and Crooke’s cell morphology
Filip Garbicz, Dawid Mehlich, Beata Rak, Emir Sajjad, Maria Maksymowicz, Wiktor Paskal, Grzegorz Zieliński, Paweł K. Włodarski
Pituitary.2017; 20(4): 450. CrossRef - Bromocriptine Induces Autophagy-Dependent Cell Death in Pituitary Adenomas
Xin Geng, Lixin Ma, Zefu Li, Zhenzhu Li, Jianmin Li, Meng Li, Qingbo Wang, Zheng Chen, Qikai Sun
World Neurosurgery.2017; 100: 407. CrossRef - Biomarkers of pituitary carcinomas
Aydin Sav, Fabio Rotondo, Luis V. Syro, Antonio Di Ieva, Michael D. Cusimano, Kalman Kovacs
Expert Review of Endocrinology & Metabolism.2016; 11(3): 253. CrossRef - MicroRNAs in the pituitary
Erica Gentilin, Ettore degli Uberti, Maria Chiara Zatelli
Best Practice & Research Clinical Endocrinology & Metabolism.2016; 30(5): 629. CrossRef - Molecular markers in pituitary tumors
Asha M. Robertson, Anthony P. Heaney
Current Opinion in Endocrinology, Diabetes & Obesity.2016; 23(4): 324. CrossRef - MicroRNA-106b promotes pituitary tumor cell proliferation and invasion through PI3K/AKT signaling pathway by targeting PTEN
Kai Zhou, Tingrong Zhang, YanDong Fan, Serick, Guojia Du, Pengfei Wu, Dangmurenjiafu Geng
Tumor Biology.2016; 37(10): 13469. CrossRef - Isolated double adrenocorticotropic hormone-secreting pituitary adenomas: A case report and review of the literature
JIUJUN PU, ZHIMING WANG, HUI ZHOU, AILING ZHONG, KAI JIN, LUNLIANG RUAN, GANG YANG
Oncology Letters.2016; 12(1): 585. CrossRef - Progress in Endocrine Neoplasia
Samuel A. Wells
Clinical Cancer Research.2016; 22(20): 4981. CrossRef - Pituitary adenomas: historical perspective, surgical management and future directions
Debebe Theodros, Mira Patel, Jacob Ruzevick, Michael Lim, Chetan Bettegowda
CNS Oncology.2015; 4(6): 411. CrossRef - Epidrug mediated re-expression of miRNA targeting the HMGA transcripts in pituitary cells
Mark O. Kitchen, Kiren Yacqub-Usman, Richard D. Emes, Alan Richardson, Richard N. Clayton, William E. Farrell
Pituitary.2015; 18(5): 674. CrossRef - Pituitary Adenoma and the Chemokine Network: A Systemic View
Fabio Grizzi, Elena Monica Borroni, Alessandro Vacchini, Dorina Qehajaj, Manuela Liguori, Sanja Stifter, Maurizio Chiriva-Internati, Antonio Di Ieva
Frontiers in Endocrinology.2015;[Epub] CrossRef - MiR-132, miR-15a and miR-16 synergistically inhibit pituitary tumor cell proliferation, invasion and migration by targeting Sox5
Wang Renjie, Liang Haiqian
Cancer Letters.2015; 356(2): 568. CrossRef - Brief Review of Articles in 'Endocrinology and Metabolism' in 2013
Won-Young Lee
Endocrinology and Metabolism.2014; 29(3): 251. CrossRef - Epigenetics of pituitary tumours
William E. Farrell
Current Opinion in Endocrinology, Diabetes & Obesity.2014; 21(4): 299. CrossRef - The Role of Genetic and Epigenetic Changes in Pituitary Tumorigenesis
Hidenori FUKUOKA, Yutaka TAKAHASHI
Neurologia medico-chirurgica.2014; 54(12): 943. CrossRef
- Plurihormonal Pituitary Neuroendocrine Tumors: Clinical Relevance of Immunohistochemical Analysis

- A Case of Adipsic Hypernatremia Associated with Anomalous Corpus Callosum in Adult with Mental Retardation.
- Boo Gyoung Kim, Ka Young Kim, Youn Jeong Park, Keun Suk Yang, Ji Hee Kim, Hee Chan Jung, Hee Chul Nam, Young Ok Kim, Yu Seon Yun
- Endocrinol Metab. 2012;27(3):232-236. Published online September 19, 2012
- DOI: https://doi.org/10.3803/EnM.2012.27.3.232
- 2,162 View
- 28 Download
- 2 Crossref
-
 Abstract
Abstract
 PDF
PDF - Adipsic hypernatremia cause chronic hyperosmolality and hypernatremia through a combination of impaired thirst and osmotically stimulated antidiuretic hormone secretion. This syndrome can be grouped together as disorders of osmoreceptor dysfunction due to the various degrees of osmoreceptor destruction related with different types of intracranial lesions around the anterior hypothalamus, consistent with the location of primary osmoreceptor cells. Adipsic hypernatremia, associated with developmental disorder of corpus callosum, is very rare. Most cases are diagnosed at infancy and early childhood; the replacement of desmopressin is necessary. Herein, we report adipsic hypernatremia associated with anomalous corpus callosum in adult with mental retardation; they were treated with only free water without desmopressin.
-
Citations
Citations to this article as recorded by- Adipsic Hypernatremia after Clipping of a Ruptured Aneurysm in the Anterior Communicating Artery: A Case Report
Won Ki Kim, Taeho Lee, Ae Jin Kim, Han Ro, Jae Hyun Chang, Hyun Hee Lee, Wookyung Chung, Ji Yong Jung
Electrolytes & Blood Pressure.2021; 19(2): 56. CrossRef - The use of diffusion tractography to characterize a corpus callosum malformation in a dog
Philippa J. Johnson, Erica F. Barry, Wen‐Ming Luh, Emma Davies
Journal of Veterinary Internal Medicine.2019; 33(2): 743. CrossRef
- Adipsic Hypernatremia after Clipping of a Ruptured Aneurysm in the Anterior Communicating Artery: A Case Report

- Expression of miRNA 146a/b, 221 and 222 in Thyroid Cancer.
- Young Suk Jo, Ihn Suk Lee, Woojeong Hong, In Sang Song, Minho Shong, Je Ryoung Kim
- J Korean Endocr Soc. 2009;24(1):17-24. Published online March 1, 2009
- DOI: https://doi.org/10.3803/jkes.2009.24.1.17
- 1,662 View
- 22 Download
- 1 Crossref
-
 Abstract
Abstract
 PDF
PDF - BACKGROUND
miRNAs can be diagnostic markers and therapeutic targets in cancers, but few studies have been conducted in thyroid cancer. We investigated the expression levels of miRNA 146a/b, 221, and 222 which are important miRNAs in papillary thyroid cancers (PTCa), and verified their impact on clinicopathological factors. METHODS: We measured the expression of pre-miRNAs 146a/b, 221, and 222 in NPA cells treated with 10% fetal bovine serum (FBS) or in HEK293T cells transfected with RET/PTC3 or BRAFV600E expression vectors. We also investigated the relationship between miRNA expression levels in thyroid cancer tissue specimens and clinicopathological parameters. RESULTS: Growth stimulation with 10% FBS induced miRNA expressions in NPA cells, and transfection of RET/PTC3 and BRAFV600E also increased the expression of these miRNAs in HEK293T cells. Most (25 cases; 50%) of PTCa showed increased expression of miRNA-146a/b and 30 cases (60%) had elevated expression of miRNA-221 and miRNA-222 compared to normal thyroid samples from the contralateral lobe. However, increased miRNA expression did not correlate with clinicopathological factors. CONCLUSION: Expression of miRNA 146a/b, 221, and 222 was increased by BRAFV600E and RET/PTC3 rearrangement and might have a role in tumorigenesis in PTCa. However, expression levels of these miRNAs did not correlate with clinicopathological parameters of patients with PTCa. -
Citations
Citations to this article as recorded by- Expression of miRNA 146a/b, 221 and 222 in Thyroid Cancer
Do Joon Park
Journal of Korean Endocrine Society.2009; 24(1): 15. CrossRef
- Expression of miRNA 146a/b, 221 and 222 in Thyroid Cancer

- Interaction of Pituitary Adenylate Cyclase-Activating Polypeptide and Angiotensin II on Aldosterone Production in Human Adrenocortical H295R Cells.
- Seong Yeon Kim, Sang Wan Kim, Young Min Cho, Do Joon Park, Chan Soo Shin, Kyung Soo Park, Bo Youn Cho, Hong Kye Lee
- J Korean Endocr Soc. 2003;18(3):272-282. Published online June 1, 2003
- 964 View
- 19 Download
-
 Abstract
Abstract
 PDF
PDF - BACKGROUND
Evidence is accumulating that aldosterone secretion can be regulated in a paracrine and/or an autocrine manner by several neuropeptides locally released within the adrenal gland. Among neuropeptides, pituitary adenylate cyclase-activating polypeptide (PACAP) is present in high concentration in the human adrenal gland. The purpose of this study was to investigate the action of PACAP and the interaction between PACAP and angiotensin II (AII), the main physiologic aldosterone secretagogue, in aldosterone production in human H295R adrenocortical cells. METHODS: H295R cells were incubated with increasing concentrations of PACAP (10(-11)M~10(-7)M) in the absence or presence of 10(-7)M AII. Aldosterone concentration in the supernatant was determined by RIA. Intracellular cAMP content was measured by RIA and total inositol phosphate (IP) production by anion exchange chromatography. Gene expression of CYP11B2 was studied by RT-PCR. RESULTS: In H295R cells, PACAP stimulated aldosterone production in a dose-dependent manner. Incubation of H295R cells with PACAP in the presence of AII significantly increased aldosterone production, compared with that of PACAP alone. PACAP dose-dependently increased cAMP production, but 10(-7)M AII had no effect on either basal or PACAP-stimulated cAMP production. Total IP production was not affected by PACAP, but was increased by 10(-7)M AII; an increase that was not further increased by addition of PACAP. RT-PCR analysis of H295R cells which were exposed to 10-7M PACAP or 10(-7)M AII showed an increase in CYP11B2 transcript signal. Induction of CYP11B2 mRNA expression in response to treatment with both PACAP and AII was significantly more than that resulting from using PACAP alone. CONCLUSION: The present study demonstrates that PACAP exerts a direct stimulatory effect on aldosterone production through induction of CYP11B2 mRNA expression by adenylate cyclase activation as the main intracellular signal pathway in H295R cells. Furthermore, there may be some additive effects between PACAP and AII on aldosterone production.


 KES
KES

 First
First Prev
Prev