Gene Expression Regulation by Agonist-Independent Constitutive Signaling of Melanocortin-1 Receptor
Article information
Abstract
Background
Melanocortin-1 receptor (Mc1r), a key signaling receptor for melanogenesis, has been reported to mediate migration of B16F10 melanoma cells. Interestingly, this activity appears to be a part of the constitutive signaling of Mc1r.
Methods
We carried out small interfering RNA-mediated knock-down of Mc1r on murine melanoma B16F10 cells and performed microarray analysis to characterize changes in the gene expression profile.
Results
We isolated 22 and four genes whose expression decreased and increased, respectively, by 2.5-fold or higher as the result of Mc1r knock-down. Several down-regulated genes have been proposed to be involved in cell migration. Among these genes are several members of the chemokine gene family.
Conclusion
We provide a gene set for further functional analyses of Mc1r. The Mc1r target genes we present may be particularly relevant for understanding the ligand-independent activity of Mc1r. Further examination of the mode of action may lead to novel strategies in regulating the migration and metastasis of melanoma cells.
INTRODUCTION
Melanocortin-1 receptor (Mc1r), a G-protein-coupled receptor expressed in melanocytes and melanoma, plays a key role in the pigmentation of hair and skin [1]. Several lines of recent evidence indicate that Mc1r also mediates cell signals regulating migration of melanoma cells. First, agouti signal protein, a well-known ligand for Mc1r has been shown to stimulate the migratory activity of melanoma cells [2]. Second, another ligand, α-MSH, inhibits migration and metastasis of melanoma cells [3,4]. Finally, we have recently reported that down-regulation of Mc1r via RNA interference inhibits both migration and metastasis of B16F10 murine melanoma cells [5]. That simple knock-down of Mc1r inhibited migration in turn suggests that constitutive signaling by Mc1r may be pro-migratory. Thus far, the promigratory signaling network down-stream of Mc1r has not been characterized at the molecular level, unlike that of promelanogenic signals. This applies to identification and functional characterization of both the cytoplasmic components of the signaling network and the ultimate targets of transcriptional activation or repression.
Here, we report gene expression profiling of highly migratory B16F10 melanoma cells following inhibition of Mc1r expression. We present several Mc1r-target genes that are potentially involved in cell migration. It appears that constitutive signaling of Mc1r positively regulates expression of several chemokines. The implications of our data are discussed.
METHODS
Cell culture
Murine melanoma cell line B16F10 was purchased from American Type Culture Collection. Cells were cultured in Dulbecco's Modification of Eagle's Medium supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin. Transwell migration assay was carried out according to our previously published protocol [5].
RNA interference
A synthetic 21-nucleotide RNA duplex specific for Mc1r (wild-type [WT]-Mc1r) and a control RNA duplex with a 5 nucleotide mismatch (mutant [MT]-Mc1r) were purchased from Dharmacon Research Inc. (Lafayette, CO, USA). The sequences are illustrated in Fig. 1A. For migration assay or microarray screen, siRNAs were used at a concentration of 50 nM. Transfection was carried out with oligofectamine (Invitrogen, Carlsbad, CA, USA) following the manufacturer's protocol. Details are available upon request.

(A) Sequences of siRNAs, wild-type melanocortin-1 receptor (WT-Mc1r), and mutant (MT)-Mc1r. (B) B16F10 cells treated with siRNAs were analyzed via transwell migration assay. Transfection of WT-Mc1r at 50 nM led to a significant reduction in migration of the cells compared to transfection of MT-Mc1r at 50 nM. (C) Confirmation of Mc1r down-regulation by reverse transcription polymerase chain reaction (RT-PCR). Two control genes (Ald1a and Ctbp1) were used to confirm the specificity of the siRNAs, and glyceraldehyde 3-phosphate dehydrogenase expression levels were used for normalization. Values represent the average of three independent real-time PCR experiments, each carried out in duplicate, and error bars represent standard deviation. aSignificant difference with a P value of 0.05.
Real-time reverse transcription polymerase chain reaction
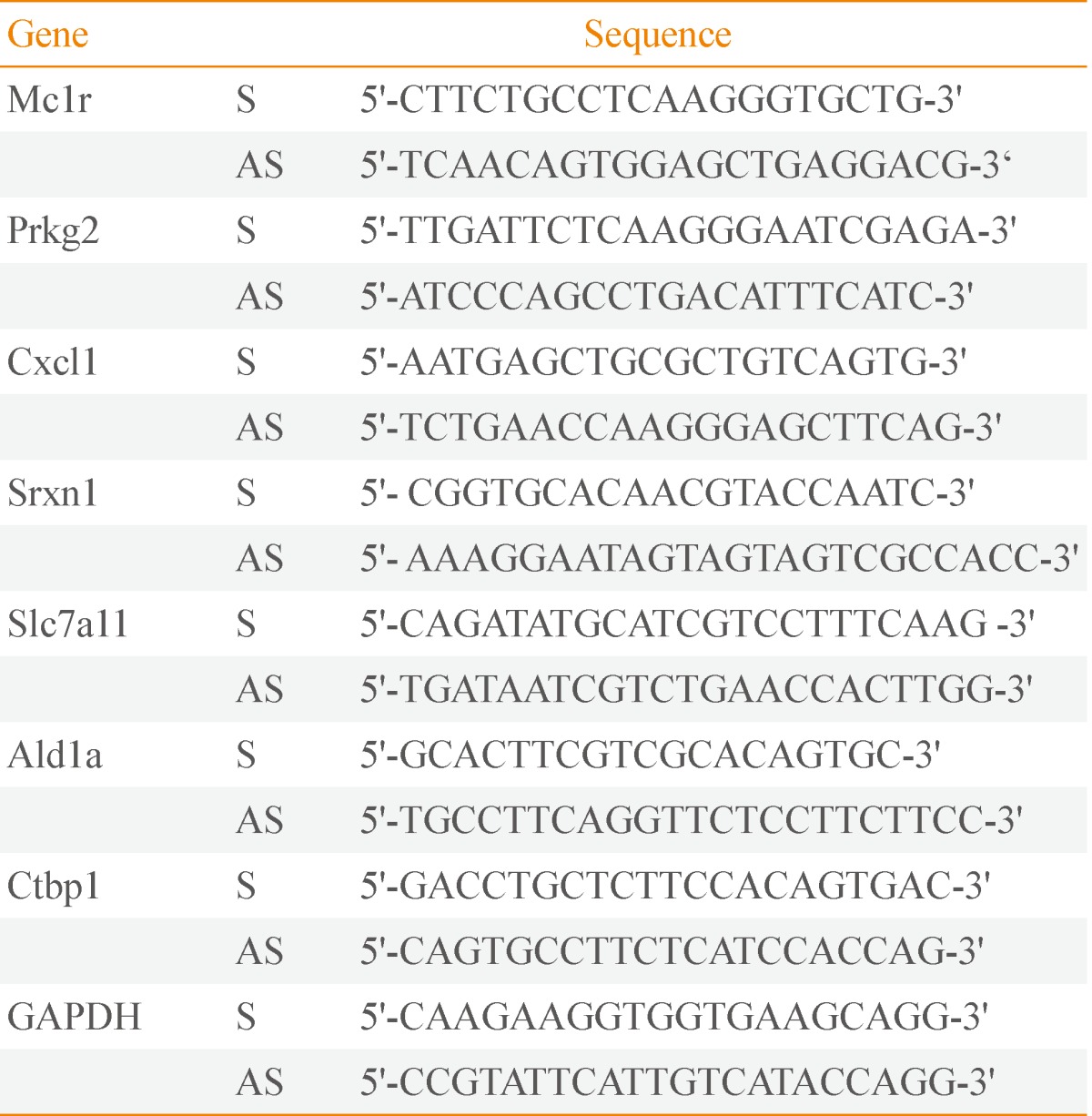
Total RNA was prepared with RNeasy Mini Kit (QIAGEN, Valencia, CA, USA) following the manufacturer's instructions. cDNA synthesis was carried out with the SuperScriptII First-Strand Synthesis System for reverse transcription polymerase chain reaction (RT-PCR, Invitrogen) using oligo-dT primers according to the manufacturer's instructions. Quantitative analyses of gene expression level were performed by real-time PCR with SYBR Green Master mix (Applied Biosystems, Foster City, CA, USA). The expression level of glyceraldehyde 3-phosphate dehydrogenase was used for normalization, and the oligonucleotide primers used are listed in Table 1.
Immunoblotting
Cells were washed twice with phosphate buffered saline and lysed in cell culture lysis buffer with protease inhibitor cocktail (Roche, Mannheim, Germany) and phenylmethylsulfonyl fluoride. Then, 20 µg of protein was separated by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis, transferred to nitrocellulose membrane, and immunoblotted with specific antibodies against mouse Srxn1 (a kind gift from professor W. Jeong at Ewha Womans University, Seoul, Korea) and β-actin (LF-PA0066, AbFRONTIER, Seoul, Korea).
Microarray screen
Total RNA was prepared with the RNeasy Mini Kit (QIAGEN) according to the manufacturer's instructions and was subsequently processed to yield biotinylated cRNA using the Ambion Illumina RNA amplification kit (Ambion, Austin, TX, USA) according to the manufacturer's instructions. The labeled cRNA preparations were applied to a MouseRef-8 v2 Expression BeadChip (Illumina, San Diego, CA, USA). Details of subsequent detection and quantitative analyses are available upon request.
RESULTS
We used B16F10 murine melanoma cells to investigate the role of Mc1r in cell migration. B16F10 cells are highly migratory and have been used to demonstrate that knock-down of Mc1r leads to inhibition of migration and metastasis [5]. In order to identify down-stream target genes of Mc1r, we carried out microarray-based gene expression profiling in combination with siRNA-mediated knock-down of Mc1r. We selected a siRNA-targeting murine Mc1r (WT-Mc1r) known to efficiently inhibit migration of B16F10 cells (Fig. 1A). A mutant siRNA with five mismatches in nucleotide sequence (MT-Mc1r) was used as the control (Fig. 1A). Migration was effectively inhibited in WT-Mc1r-treated cells compared to MT-Mc1r-treated cells (Fig. 1B). RT-PCR assay indicated that WT-Mc1r reduced the mRNA level to about 50% of the level seen in MT-Mc1r treatment (Fig. 1C). This change in expression was not seen with the two negative control genes, Ald1a and Ctbp1, consistent with the specific activity of WT-Mc1r (Fig. 1C). Microarray data were in agreement with RT-PCR results. Mc1r was down-regulated 2.15-fold by WT-Mc1r siRNA while Ald1a and Ctbp1 were not affected (data not shown).
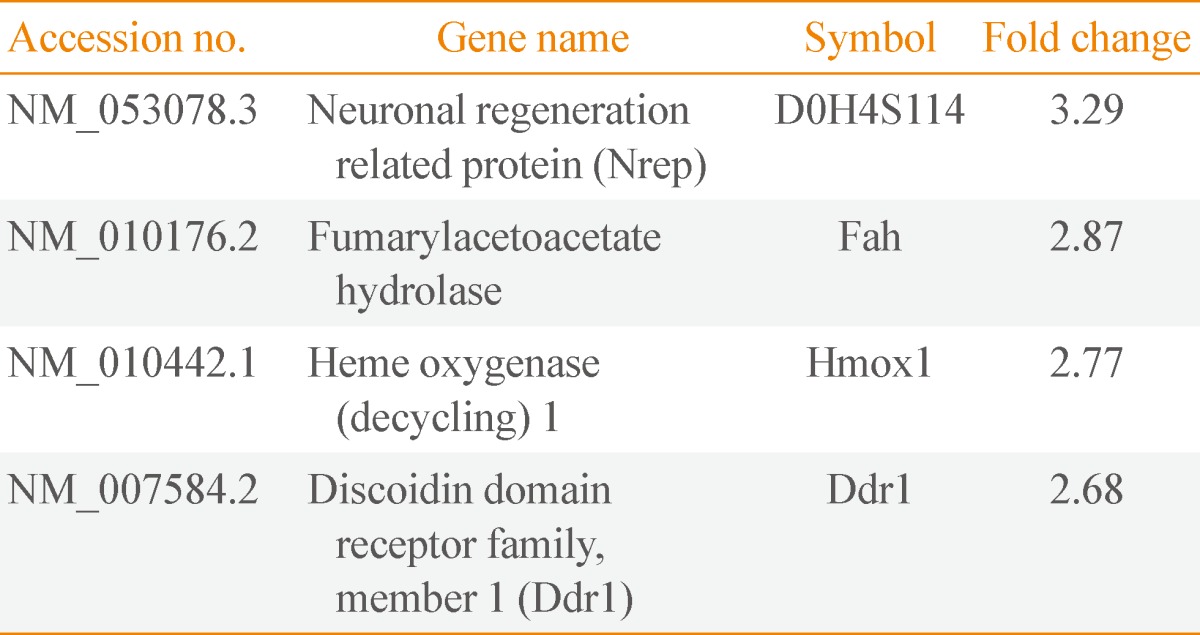
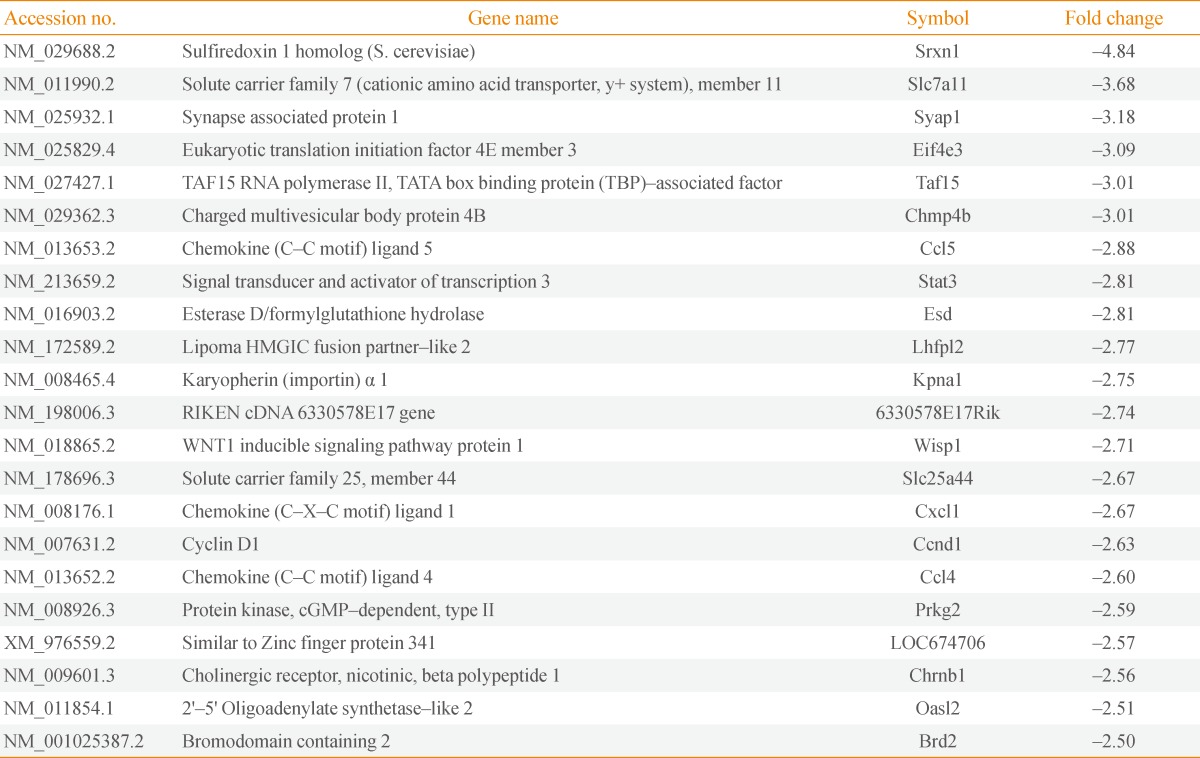
Tables 2, 3 show genes whose expression levels were up- and down-regulated, respectively, by more than 2.5-fold after averaging values from duplicate microarray assays. We confirmed the microarray data by RT-PCR on a subset of genes down-regulated by WT-Mc1r (Fig. 2A). We also examined the down-regulation of sulfiredoxin 1 (Srxn1), which exhibited the largest reduction in expression, by immunoblotting. Consistent with the results from RT-PCR, Srxn1 protein demonstrated visible down-regulation by WT-Mc1r (Fig. 2B).
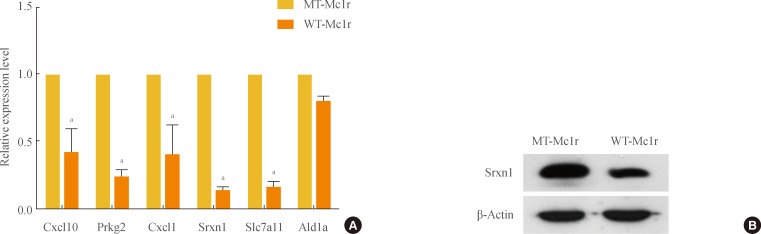
(A) Confirmation of microarray data by reverse transcription polymerase chain reaction (RT-PCR). A subset of genes that showed down-regulation of 2.5-fold or higher by wild-type melanocortin-1 receptor (WT-Mc1r) compared to mutant (MT)-Mc1r in microarray assay and a nontarget gene (Ald1a) whose expression level was unchanged were used to validate the results from the microarray assay. Glyceraldehyde 3-phosphate dehydrogenase expression levels were used for normalization. Values represent the average of three independent real-time PCR experiments, each carried out in duplicate, and error bars represent standard deviation. (B) Immunoblotting for Srxn1 shows down-regulation by WT-Mc1r. β-Actin was used as the loading control. aSignificant difference with a P value of 0.05.
Srxn1 is a member of an antioxidant protein family containing a ParB-like nuclease domain. At least two recent studies reported that Srxn1 promotes cell migration [6,7]. Another target gene, Stat3, a member of the signal transducer and activator of transcription family of transcription factors, is also known to promote cell motility via multiple mechanisms [8,9].
Notable among the genes down-regulated by WT-Mc1r are three chemokines, Ccl5, Cxcl1, and Ccl4. Ccl5, also known as RANTES, promotes the migration of multiple cell types [10,11,12] and is expressed in multiple metastatic human melanomas [13]. Cxcl1, also known as melanoma growth-stimulating activity-α, is involved in melanoma pathogenesis and has been shown to promote the migration of multiple lines of human melanoma cells [14,15]. Ccl4, also known as macrophage inflammatory protein-1β, likewise has been reported to increase the migratory activity of human uveal melanoma cells [16].
DISCUSSION
In this report, we describe the changes in gene expression following knock-down of Mc1r by RNA interference. The genes thus identified represent regulatory targets of the ligand-independent signaling of Mc1r. To the best of our knowledge, this is the first study that identifies such a group of genes. At least five genes among those down-regulated have been reported to promote cell migration in various contexts. The most notable are the three chemokines, Ccl5, Cxcl1, and Ccl4. Although their expression and/or promigratory effect in melanoma cells have been reported previously, this is the first time that a regulatory link has been established with Mc1r signaling.
Future experiments should examine whether these genes are functional in melanoma migration. The promigratory activities of Srxn1 and Stat3 have yet to be confirmed in melanoma. It will also be necessary to directly test if these chemokines promote migration of melanoma and, if so, whether they function as autocrines. Inhibitors of Srxn1antioxidant activity or of Stat3 activation and nuclear localization could also be developed and used as repressors of migration, and possibly of metastasis. For chemokines, competitive inhibitors for receptor binding or neutralizing antibodies can be tested for their effects on cell migration. Detailed knowledge of the Mc1r signaling network and its promotion of melanoma migration, including down-stream effector genes, could provide the opportunity for development of novel therapeutic strategies for melanoma.
ACKNOWLEDGMENTS
This research was supported by a grant (2010K000803) from the Brain Research Center of the 21st Century Frontier Research Program and by the "Systems biology infrastructure establishment grant" provided by the Gwangju Institute of Science & Technology in 2008 through the Ewha Research Center for Systems Biology (ERCSB).
Notes
No potential conflict of interest relevant to this article was reported.