Regulation of Adipocyte Differentiation via MicroRNAs
Article information
Abstract
Adipocyte differentiation, termed adipogenesis, is a complicated process in which pluripotent mesenchymal stem cells differentiate into mature adipocytes. The process of adipocyte differentiation is tightly regulated by a number of transcription factors, hormones and signaling pathway molecules. Recent studies have demonstrated that microRNAs, which belong to small noncoding RNA species, are also involved in adipocyte differentiation. In vivo and in vitro studies have revealed that various microRNAs affect adipogenesis by targeting several adipogenic transcription factors and key signaling molecules. In this review, we will summarize the roles of microRNAs in adipogenesis and their target genes associated with each stage of adipocyte differentiation.
INTRODUCTION
Adipose tissue plays key roles in energy storage, regulation of body temperature and absorption from mechanical collision. In a state of obesity, adipose tissue expands due to an increase in adipocyte size (hypertrophy) and/or number (hyperplasia), thereby inducing dysregulation of glucose and lipid metabolism. As a result, increased adipose tissue destroys whole body energy balance and enhances the risk of insulin resistance, hypertension, and dyslipidemia. Therefore, proper understanding of adipocyte differentiation would provide valuable information for designing comprehensive and effective therapeutic strategies against obesity.
Adipocyte differentiation occurs in several stages, involves many signaling pathways, and progress depends on various stimuli such as nutrients and hormones. For example, adipogenesis is tightly controlled by a cascade of several transcription factors such as CCAAT/enhancer-binding proteins (C/EBPs) and peroxisome proliferator-activated receptor γ (PPARγ). Additionally, several signaling molecules, including wingless and INT-1 proteins (Wnts) and insulin, modulate adipogenesis. It has also been demonstrated that microRNAs are involved in adipocyte differentiation [1].
MicroRNAs (miRNA) are 19 to 22 nucleotide fragments of noncoding RNA that play important roles in various cellular processes through posttranscriptional regulation of target genes. As many groups have reported that miRNAs actively participate in cell proliferation and differentiation [2,3,4], knowledge is constantly increasing related to miRNAs involvement in adipogenesis and fat metabolism. Although several reviews have recently been published on related topics [1,5,6], this review article will specifically describe the miRNA-mediated regulatory mechanism in adipocyte differentiation, particularly by updating and integrating the most recent information.
MOLECULAR EVENTS DURING ADIPOCYTE DIFFERENTIATION
In vitro studies have suggested that the progression of adipocyte differentiation has, at least, two key steps: commitment and differentiation. Adipocytes are derived from pluripotent mesenchymal stem cells (MSCs) that have the capacity to develop into several cell types, including adipocytes and osteoblasts. Commitment or determination of MSCs' fate to differentiate into preadipocytes is caused by adipogenic differentiation signaling cues that have not yet been indentified [7]. Following this step, committed MSCs are specified for an adipogenic lineage and often lose their ability to differentiate into other cell lineages.
In the differentiation step, committed preadipocytes derived from MSCs (e.g., 3T3-L1 cells) are differentiated into adipocytes after exposure to hormone cocktails such as insulin, dexamethasone and cyclic adenosine monophosphate (cAMP) activators [8]. Contact with these chemicals induces G1 phase-arrested 3T3-L1 cells to synchronously undergo, on average, two cycles of cell division, so called mitotic clonal expansion. During the cell cycle, clonal expansion is regulated by the Rb-E2F pathway, which is responsible for the G1-to-S transition. Rb inhibits the cell cycle by binding to, and repressing, the transcriptional activity of E2F. Upon hyperphosphorylation of Rb by cyclin-dependent kinases, E2F is released and promotes transcriptional activation of genes that encode cell-cycle regulators required for S phase entry; a process that initiates clonal expansion [9]. In growth-arrested preadipocytes, there are significant levels of Rb family p130 (pRB/p130). Inactivation of Rb2/p130 by phosphorylation enables clonal expansion. When cells exit the cell cycle, E2F loses its activity and terminal differentiation is initiated [10].
Several signaling pathways highlight molecules such as bone morphogenic protein (BMP) and Wnt, which have been shown to be key molecules in the regulation of MSC commitment to adipocyte lineage and the differentiation of a subset of adipocytes. BMPs belong to the transforming growth factor β (TGF-β) family of growth factors, which consists of 14 family members. BMP-2 and BMP-4 have been implicated in adipogenesis and are thought to promote commitment of cells to adipogenic lineages [11,12,13,14]. The positive role of BMP-4 in adipocyte commitment has been demonstrated with several established cell lines. In C3H10T1/2 cells, exogenous BMP-4 activation induces potent adipocyte differentiation. In addition, a committed preadipocyte A33 cell line derived from C3H10T1/2 stem cells expresses and secretes BMP-4 at the same time point when exogenous BMP-4 is added to C3H10T1/2 cells for adipogenic differentiation. Furthermore, exposure of A33 cells to noggin, a naturally occurring BMP-4-binding antagonist, during this critical time window blocks subsequent differentiation [11]. The effect of BMP-2 is more complex. BMP-2 can enhance adipogenesis of C3H10T1/2 cells at low concentrations, but stimulates chondrocyte and osteoblast development at higher concentrations [14]. In preadipocytes, BMPs activate Sma and Mad related protein (Smad) signaling and regulate many target genes including cytoskeleton-associated proteins [12]. BMPs are also known as powerful cytokines that induce bone and cartilage formation. BMP-Smad signaling in this developmental context can activate runt-related transcription factor 2 (Runx2), osterix, Dlx5/6, and Sox9, which are essential transcription factors for osteogenesis and chondrogenesis [13].
In addition to BMPs, the TGF-β superfamily member, TGF-β, is also involved in adipogenesis. In general, TGF-β signals through two types of transmembrane serine/threonine kinase receptors, type I and type II TGF-β receptors, and signaling effector Smads. Activation of Smad2 or Smad3 by TGF-β receptors results in heterodimerization with Smad4 and stimulates nuclear translocation of Smad complexes. In the nucleus, Smad proteins regulate transcription by binding to DNA and interacting with other transcription factors. During adipogenesis, TGF-β phosphorylates only Smad3, which then binds to C/EBPs and inhibits their transcriptional activity, including the ability to transactivate PPARγ [15]. Consistently, it has been demonstrated that TGF-β1 inhibits the early stages of 3T3-L1 differentiation [16] by promoting the proliferation of progenitor cells and hampering lipid accumulation [17]. Moreover, transgenic overexpression of TGF-β in adipose tissue inhibits differentiation in vivo [18].
The Wnt family is made up of secreted glycoproteins that influence cell fate and development. Wnt proteins bind to frizzled receptors to stimulate signaling cascades through β-catenin-dependent (Wnt/β-catenin) and -independent pathways. The Wnt/β-catenin signaling pathway is often activated in preadipocytes and expression of Wnts declines after induction of differentiation [19,20]. It has been suggested that Wnt/β-catenin signaling enhances proliferation during commitment and mitotic clonal expansion [20]. Activated Wnt/β-catenin signaling enables the lymphoid-enhancer-binding factor/T-cell-specific transcription factor (LEF/TCF) family of transcription factors to activate Wnt target genes. In late stages of adipogenesis, Wnt/β-catenin signaling inhibits adipogenesis; however little is known about how Wnt inhibits adipogenesis through TCF/LEF [21].
Many signaling pathways influence adipocyte differentiation. For instance, mitogen-activated protein kinase (MAPK) pathways may enhance adipogenesis through extracellular signal-regulated kinases (ERKs) and p38. Although it has been suggested that activation of ERK1 positively regulates adipogenesis during clonal expansion, activity has to be reduced after proliferation in order for adipogenesis to proceed [22]. Among various hormones, insulin plays a key role in adipogenesis. Insulin functions predominantly through insulin growth factor-1 receptor signaling, and the downstream signaling involves insulin receptor substrate (IRS), phosphoinositide 3-kinase (PI3K), PDK1, and AKT/protein kinase B (PKB) signaling. Insulin signaling can be transmitted to the adipogenic cascades in several different ways. IRS signaling is known to promote cAMP response element-binding (CREB) phosphorylation, thereby influencing adipogenesis [23]. Another way for insulin to deliver its signal is through inhibition of antiadipogenic forkhead box protein O (FOXO) transcription factors after AKT/PKB-mediated phosphorylation [24].
Induction of adipogenesis by differentiation cocktails immediately phosphorylates and activates CREB, which in turn transcriptionally activates the C/EBPβ gene. Elevated C/EBPβ is somewhat inactive until it acquires DNA-binding activity through a series of phosphorylaton events by MAPK and GSK3β. Early activation of C/EBPβ, together with C/EBPδ, promotes gene expression of C/EBPα and PPARγ through C/EBP regulatory elements in their proximal promoters. C/EBPα induces adipogenic genes, and in vivo studies indicate an important role for this factor in the development of adipose tissue. PPARγ is the 'master regulator' of adipogenesis and is a necessary and sufficient factor for adipogenesis. Even though the C/EBPs are considered to be important transcription factors in adipogenesis, they cannot function efficiently without PPARγ [25]. PPARγ and C/EBPα coordinately control expression of a large group of genes that are required for adipocyte phenotypes. During adipogenesis, PPARγ and C/EBPα positively cross-activate each other through their respective C/EBP regulatory elements [25,26].
Other key transcriptional factors are the Kruppel-like factors (KLFs). The KLFs are a large family of C2H2 zinc-finger proteins that regulate apoptosis, proliferation, and differentiation. This family of transcription factors contains both repressors and activators of transcription. Seven of the 17 known KLF proteins have been shown to be involved in different phases of adipocyte differentiation (i.e., KLF2 and 3 inhibit differentiation, whereas KLF4, 5, 6, 9, and 15 stimulate differentiation) [27].
ROLES OF miRNAs IN ADIPOGENESIS
A number of miRNAs and their targets have been implicated in adipogenesis. As summarized in Table 1 [1,2,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68], some appear to enhance adipocyte differentiation while others inhibit adipogenesis in various model systems. An integrative model displaying the function of each miRNA in the progression of adipogenesis is depicted in Fig. 1.
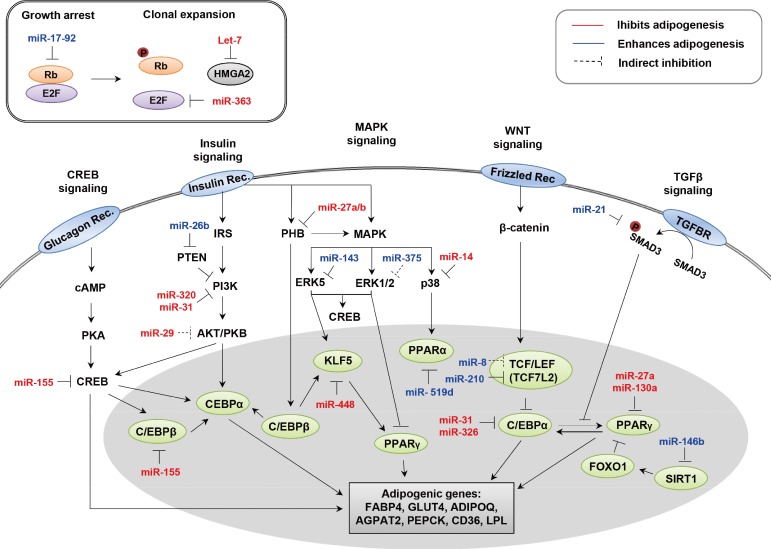
Signals and microRNAs involved in adipogenesis. HMGA2, high mobility group AT-hook2; CREB, cAMP response element-binding; MAPK, mitogen-activated protein kinase; WNT, wingless and INT-1; TGF-β, transforming growth factor β; TGFBR, TGF-β receptor; IRS, insulin receptor substrate; PHB, prohibitin; SMAD3, Sma and Mad related protein 3; cAMP, cyclic adenosine monophosphate; PTEN, phosphatase and tensin homolog gene; PI3K, phosphoinositide 3-kinase; ERK, extracellular signal-regulated kinase; PKA, protein kinase A; PKB, protein kinase B; KLF, Kruppel-like factor; PPAR, peroxisome proliferator-activated receptor; TCF, T-cell-specific transcription factor; LEF, lymphoid-enhancer-binding factor; C/EBP, CCAAT/enhancer-binding protein; FOXO, forkhead box protein O; SIRT1, sirtuin 1; FABP4, fatty acid binding protein 4; GLUT4, glucose transporter type 4; ADIPOQ, adiponectin; AGPAT2, 1-acyl-sn-gylcerol-3-phosphate acyltransferase beta; PEPCK, phosphoenolpyruvate carboxykinase; LPL, lipoprotein lipase.
LINEAGE DETERMINATION (COMMITMENT)
Adipocytes and osteoblasts originate from common MSCs. The differentiation of MSCs into adipocytes and osteoblasts is not only dependent on the mechanisms that determine a specific cell lineage, but also on the mechanisms that suppress the development of other lineages [69]. In mesenchymal progenitors, the selection between adipogenesis and osteogenesis is influenced by reciprocal regulation of different intracellular signals and transcription factors. Furthermore, previous studies have suggested that miRNAs are involved in both the lineage fate of MSCs and osteoblast differentiation [70].
PPARγ regulates the whole process of adipogenesis including lineage commitment and differentiation. Thus, it appears that miRNAs specifically targeting PPARγ would also be modulators of adipogenesis. Several computational prediction programs have proposed that miR-27a and miR-130a may target PPARγ 3'-untranslated region (UTR) in a sequence-specific manner. In accordance with those predictions, it has been reported that both miR-27a and miR-130a indeed suppress adipocyte differentiation through PPARγ downregulation [28,29]. In 3T3-L1 cells, the levels of miR-27a and miR-130a are gradually decreased during adipogenesis, which is inversely correlated with expression levels of PPARγ. Furthermore, overexpression of miR-27a and miR-130a evidently suppresses adipocyte differentiation, concomitantly with PPARγ protein expression. These findings have suggested that both miR-27a and miR-130a negatively regulate PPARγ expression, leading to the repression of adipocyte differentiation. However, the roles of miR-27a and miR-130a in osteogenesis have not been elucidated. Thus, examining their roles in osteogenesis may provide insight into the physiological roles of miRNAs in the differentiation of MSCs to osteoblasts in bone tissue for therapeutic purposes.
To investigate whether miR-27a and miR-130a may regulate osteogenesis, isolated primary bone marrow cells (PBMCs) were retrovirally infected with miR-27a and miR-130a (Fig. 2A, B). After infection, PBMCs were maintained under osteogenic differentiation conditions. Alkaline phosphatase (ALP) staining showed that osteoblast differentiation was not influenced by miR-27a or miR-130a (Fig. 2C). Further, in PBMCs overexpressing miR-27a and miR-130a, mRNA expression levels of osteogenic marker genes such as osteopontin and osteocalcin were not systematically changed (Fig. 2D). These results reveal that miR-27a and miR-130a do not have significant effects on osteogenesis, implying that miR-27a and miR-130a may selectively affect adipogenesis, not osteogenesis.
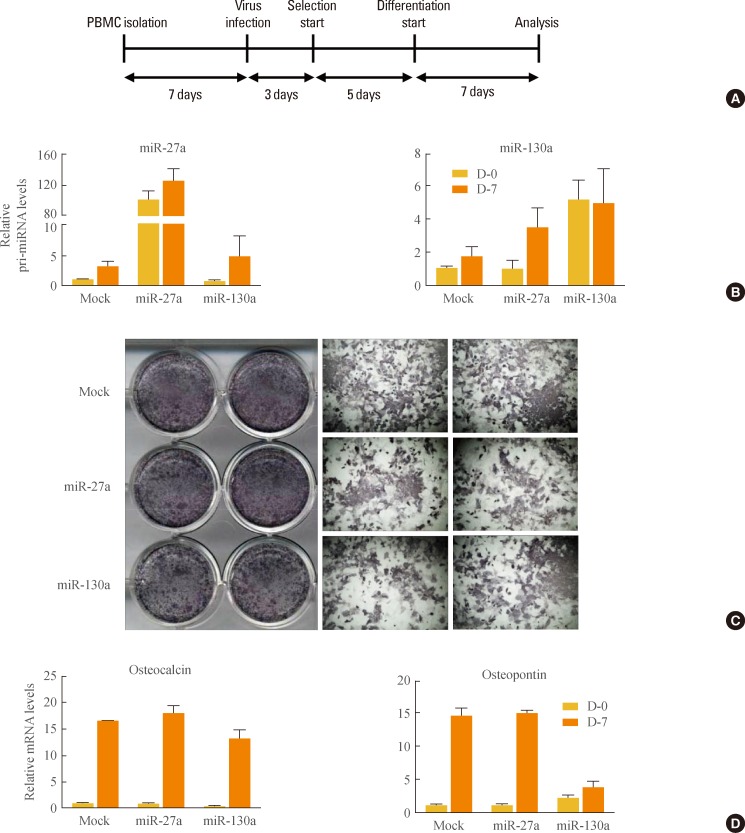
Regulation of osteogenesis by microRNA (miR)-27a and miR-130a. (A) Experimental scheme. primary bone marrow cell (PBMCs) were isolated from the femur and tibia of 4-week-old C56BL/6J mice. After 7 days, PBMCs were infected with retrovirus containing mock, full-length of pri-miR-27a or pri-miR-130a. Retrovirus-infected PBMCs were selected with puromycin for 5 days and differentiated for an additional 7 days. For osteoblast differentiation, PBMCs were cultured in α-modified essential medium with 10% fetal bovine serum, 50 µM ascorbic acid, and 10 mM β-glycerophosphate. All experiments with mice were approved by the Institute of Laboratory Animal Resources at Seoul National University. (B) Overexpression of miR-27a or miR-130a. The relative amounts of pri-miR-27a and pri-miR-130a against cyclophilin were measured with quantitative reverse transcription polymerase chain reaction (q-RT-PCR). Results are expressed as mean±standard error of mean (SEM). (C) Microscopic view of differentiated PBMCs after alkaline phosphatase (ALP) staining. Differentiated osteoblasts were monitored by ALP staining to reveal ALP-positive cells. (D) Expression of osteogenic marker genes. The relative mRNA levels of osteocalcin and osteopontin were measured by q-RT-PCR. Results are expressed as mean±SEM.
There are other known miRNAs that can control lineage determination. miR-124 is gradually upregulated during adipocyte differentiation of human bone marrow-derived mesenchymal stem cells (hMSCs). In 3T3-L1 cells, miR-124 overexpression promotes adipocyte differentiation via the suppression of Dlx5, accompanied by elevated fatty acid binding protein 4 (FABP4). Dlx5 is known as a pro-osteogenic transcription factor that determines cell fate in MSCs [71]. Thus, miR-124 has a proadipogenic effect by targeting Dlx5 [30]. Recent studies have demonstrated that miR-204, and its homolog miR-211, promote the induction of bone marrow stromal cell differentiation into adipocytes [31]. As shown by reporter assays, miR-211 directly targets 3'UTR of Runx2, which is a key transcription factor for osteoblast differentiation. Ectopic expression of miR-204 decreased Runx2 protein levels, whereas miR-204 knockdown significantly increased Runx2 protein levels. These data imply that miR-204 would suppress the action of Runx2 in osteoblast differentiation and enhance adipocyte differentiation.
miRNAs REGULATING MITOTIC CLONAL EXPANSION
Some miRNAs are involved in adipogenesis by regulating the RB-E2F pathway that controls mitotic clonal expansion. The miR-17-92 cluster has been reported to be upregulated during the clonal expansion stage of adipocyte differentiation, positively regulating adipogenesis [32]. Overexpression of miR-17-92 accelerates adipocyte differentiation and directly represses RB family Rb2/p130. On the contrary, miR-363 inhibits adipocyte differentiation by targeting E2F family member, E2F3 [72]. miR-363 overexpression downregulates expression levels of C/EBPα and PPARγ.
In addition, let-7 negatively regulates adipogenesis by regulating the expression of high mobility group AT-hook2 (HMGA-2) [33]. Ectopic introduction of let-7 in 3T3-L1 and 3T3-F442A cells decreases mRNA levels of mature adipocyte markers such as FABP4 and PPARγ. Among several genes that have been previously shown to be targets of let-7, the most affected in 3T3-L1 is HMGA2, a transcription factor. Expression of HMGA2 is high during clonal expansion but turns off at terminal differentiation. HMGA2-deficient mice exhibit a reduction in adipose tissue [73], whereas overexpression in transgenic mice leads to an increase in fatty tissue [74]. This result depicts let-7 as an anti-adipogenic regulator that regulates the transition from clonal expansion to terminal differentiation.
ANTIADIPOGENIC miRNAs
Some miRNAs control adipogenesis by blocking signal transduction pathways such as MAPK. miR-14 has a suppressive effect on fat metabolism by targeting p38 and MAPK in drosophila [34]. In flies, deletion of miR-14 increases the number of lipid droplets and the concentration of triacylglycerol. Similarly, miR-27a and miR-27b are involved in adipocyte differentiation. Computational prediction and luciferase assays have shown that miR-27a and miR-27b target prohibitin (PHB) [75], which is highly expressed in cells that rely heavily on mitochondrial function and has recently been implicated in adipogenesis [76]. In 3T3-L1 cells, overexpression of PHB inhibits insulin-induced adipogenesis, whereas in the absence of insulin, PHB facilitates adipogenesis through upregulation of MAPK/ERK signaling. PHB silencing by transfection with miR-27 impairs adipocyte differentiation in adipose tissue-derived stem cells (ASC). Likewise, protein levels of PHB, C/EBPβ, PPARγ, and aP2 are reduced in miR-27a- or miR-27b transfected ASCs. Moreover, ectopic expression of miR-27a or miR-27b attenuates lipid accumulation [35]. Considering all of these factors, miR-27a and miR-27b are therefore negative modulators of adipocyte differentiation by suppressing PHB.
Many miRNAs have been reported to negatively regulate adipocyte differentiation by targeting insulin signaling and C/EBPs. In 3T3-L1 adipocytes, miR-320 and miR-29 decrease insulin activity by targeting PI3K directly [36] and AKT indirectly [37], respectively. However, it is unclear whether they can influence adipogenesis. Tang et al. [38] identified significant downregulation of miR-31 and miR-326 during adipogenesis of ADSCs. In silico prediction of target genes suggested that miR-31 may target phosphoinositide-3-kinase, class 2, alpha polypeptide (PIK3C2A) and C/EBPα, while miR-326 targets Ras association domain family member 1 (RASSF1) and AP2 associated kinase 1 (AAK1) [38]. In addition, expression of these predicted target genes is correlated with adipogenic differentiation. Another study has demonstrated that C/EBPα protein levels are downregulated by miR-31 expression in rats [39]. A recent report also demonstrated that miR-155 is an inhibitor of adipocyte differentiation. Ectopic expression of miR-155 decreased lipid accumulation as assessed by oil-red O staining in 3T3-L1 cells [40] and adipose tissue-derived preadipocytes [77]. Overexpression of miR-155 is associated with reduced protein levels of CREB and C/EBPβ, which are essential regulators during the early stages of terminal differentiation.
Transcriptional or posttranscriptional processes related to adipogenesis are also controlled by many miRNAs. A bioinformatics approach has indicated that miR-448 targets KLF5 [41], and miR-448 is considered to be an inhibitor of adipocyte differentiation. Overexpression of miR-448 reduces KLF5 and inhibits adipocyte differentiation. mRNA levels of adipogenic marker genes such as C/EBPα and PPARγ, which are the targets of KLF5, are regulated by miR-448. In addition, miR-448 knockdown induces elevation of triglyceride concentration and upregulation of KLF5 expression, thereby promoting differentiation of 3T3-L1 preadipocytes. In 3T3-L1 cells, miR-224-5p can suppress adipocyte differentiation by targeting early growth response 2 (EGR2, also known as Krox20) [78]. EGR2 is a transcription factor that functions during early stages of adipocyte differentiation and enhances adipogenesis through C/EBPβ-dependent and -independent mechanisms. miR-224-5p is able to regulate fatty acid metabolism by directly targeting acyl-CoA synthetase long chain family member 4 (ACSL4). miR-138 has been implicated as an inhibitor of adipocyte differentiation through posttranscriptional regulation of EP300 interacting inhibitor of differentiation 1 (EID-1), which can promote adipocyte differentiation [42]. The role of miR-138 in human multipotent MSCs has been demonstrated by gain of function experiments. When miR-138 is overexpressed in hMSCs, lipid droplets are significantly reduced and the expression levels of C/EBPα and PPARγ decrease.
PROADIPOGENIC miRNAs
It has been reported that several miRNAs may promote adipocyte differentiation by targeting MAPK signaling. In 3T3-L1 cells, expression of miR-143 increases during adipogenesis, and thereby stimulates adipocyte differentiation through inhibition of ERK5 via binding to the 3'-UTR of ERK5 mRNA [43]. Similarly, miR-375 overexpression increases the mRNA expression levels of C/EBPα, PPARγ, and adipocyte FABP (aP2) [36]. The number of lipid droplets is also increased by ectopic expression of miR-375 in 3T3-L1 cells. Furthermore, induction of miR-375 represses phosphorylation levels of ERK1/2. In 3T3-L1 cells, miR-375 knockdown elevates the phosphorylation level of ERK1/2 and decreases mRNA expression levels of C/EBPα, PPARγ, and aP2.
Recent findings have suggested that miR-26b inhibits adipogenic differentiation in human preadipocytes [3]. Bioinformatics approaches and luciferase assays have revealed that the 3'UTR of the phosphatase and tensin homolog gene is a target of miR-26b. miR-26b overexpression enforces downregulation of adipogenic marker genes including aP2, C/EBPα and PPARγ. In addition, the abundance of lipid droplets and the triglyceride concentration are lowered in miR-26b-overexpressing cells. However, knockdown of miR-26b enhances adipocyte differentiation, accompanied with an increase in both adipocyte-specific marker genes and lipid droplets.
Kennell et al. [44] have reported that miR-8 can inhibit the evolutionarily-conserved Wnt/Wingless pathway in drosophila. This regulation has been recapitulated with ST2 marrow stromal cells during adipogenesis. Overexpression of the miR-8 family (miR-200c-141 and miR-200b-200a-429 clusters) increases adipogenesis, FABP4, and lipid accumulation [44]. miR-8 indirectly targets the TCF transcription factor in Wnt signaling and directly targets CG32767, a zinc finger protein identified as a positive regulator of Wnt signaling in drosophila. Furthermore, miR-210 has been reported as a proadipogenic miRNA [79]. In 3T3-L1 cells, lentiviral overexpression of miR-210 enhances adipocyte differentiation, whereas inhibition of miR-210 suppresses expression of adipogenic marker genes. miR-210 enhances adipogenesis by inhibiting TCF7L2, which is a key transcription factor in Wnt signaling.
According to the microarray results, the expression of miR-21 changes during adipocyte differentiation [80]. Oil-red O staining has demonstrated that miR-21 overexpression increases the abundance of lipid droplets and the size of adipocytes. In 3T3-L1 cells, miR-21 stimulates an increase in mRNA and protein levels of adiponectin. In addition, regulation of human ASC (hASC) differentiation by miR-21 is mediated by direct inhibition of TGF-β receptor 2 (TGFBR2) expression, which is closely related to the TGF-β signaling pathway. Recent findings suggest that miR-21 may enhance adipocyte differentiation of hASCs via inhibition of TGFBR2 [2]. miR-21 is known to have an adipogenic effect by targeting TGFBR2 in TGF-β signaling.
In addition, miR-146b has been identified as a positive modulator of adipocyte differentiation via suppression of sirtuin 1 (SIRT1) [81]. SIRT1, a target of miR-146b, works as an inhibitor of adipocyte differentiation by inducing deacetylation of FOXO1. Expression patterns of miR-146b change significantly during adipocyte differentiation. In 3T3-L1 cells, ectopic expression of miR-146b increases expression of C/EBPα, PPARγ, and aP2. Furthermore, knockdown of miR-146b significantly decreases expression of adipogenic markers, body weight and visceral fat. Interestingly, miR-146b is highly expressed in the adipose tissue of obese mouse models such as DIO, diet-induced obesity, ob/ob, and db/db. This finding has implicated miR-146b as a positive regulator of accelerated adipocyte differentiation through modulation of SIRT1.
It has also been shown that miR-519d is downregulated in obese individuals. In human subcutaneous adipose tissue, miR-519d overexpression results in an increase in adipocyte differentiation by targeting PPARα [45]. PPARα plays a crucial role in fatty acid oxidation in the liver, muscles and heart. Therefore, it is likely that miR-519d enhances adipogenesis by inhibiting fat burning through the downregulation of PPARα.
Some miRNAs exhibit clear association with adipogenesis although their target genes or processes are unknown. For example, gain of function experiments have revealed that overexpression of miR-103 in preadipocytes upregulates expression of adipogenic genes such as PPARγ. Several miR-103 studies have been reported; however, there are inconsistencies between the miRNA profiles of mice and humans [46]. Thus, further research is necessary to delineate the role of miR-103 in adipocyte differentiation. miR-378 is also involved in adipocyte development, and is highly induced during adipocyte differentiation of ST2 cells [82]. Overexpression of miR-378 increases the size of lipid droplets and seems to promote the transcriptional activity of C/EBPα and C/EBPβ. Conversely, knockdown of miR-378 decreases the rate of triglyceride accumulation in ST2 cells. miR-132 has been identified as a biomarker of obesity.
Numerous studies have revealed that miRNAs are associated with key metabolic parameters in obese patients. Microarray expression profiles have shown different circulating levels of miR-132 between obese and nonobese omental fat. Analysis of miR-132 has revealed that altered expression of miR-132 in blood is correlated with body mass index, fasting glucose, and glycosylated hemoglobin [6]. Among the targets of miR-132, CREB has been shown to be involved in regulation of appetite and glucose level. Thus, miR-132 expression may contribute to regulation of metabolic actions including energy homeostasis.
CONCLUSIONS
Because obesity-related hyperplasia and hypertrophy are often associated with various metabolic disorders, detailed under standings of the molecular events regulating adipogenesis and lipid metabolism are important. Emerging evidence shows that miRNAs may directly or indirectly modulate adipocyte different iation [6]. The regulation of adipogenesis by miRNAs includes altering the expression of genes associated with each stage of adipocyte differentiation. In this regard, miRNAs can act on adipogenic factors in either positive or negative manners. Moreover, recent findings have proposed the idea that certain miRNAs could be biomarkers of obesity. However, many challenges remain. For example, a complex redundancy of miRNAs and target interactions may exist during the com plicated processes of adipogenesis, and the nature of these interactions has not been fully characterized. Therefore, further investigations are definitely required to elucidate the precise roles of these miRNAs and their regulatory mechanisms in adipogenesis. In conclusion, more research concerning the miRNAs involved in adipocyte differentiation would provide pathophysiological roles for these miRNAs and novel insight into obesity and its related metabolic diseases.
ACKNOWLEDGMENTS
This work was supported by the National Creative Research Initiative Program (2011-0018312), funded by the Ministry of Education, Science and Technology (MEST).
Notes
No potential conflict of interest relevant to this article was reported.
